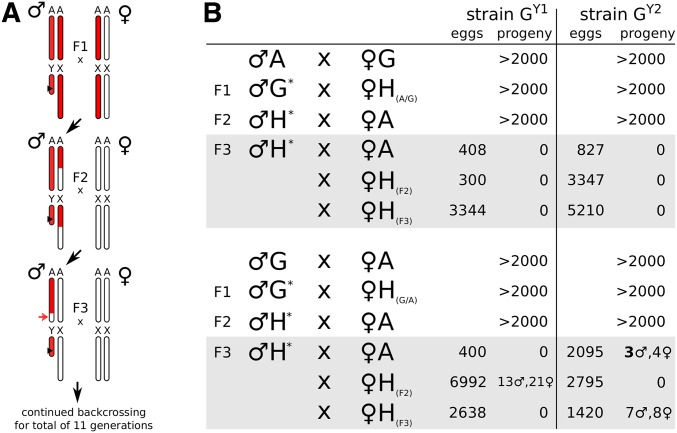
Figure 1.
F0 × F1 hybrid crossing scheme and resulting progeny. (A) Crossing scheme indicating the A. gambiae (red) or A. arabiensis (white) genomic contributions in generations F1–F3, with autosomes represented as a single pair labeled A. In the F1 cross, A. gambiae males of strains GY1 or GY2 are crossed to hybrid F1 females. Each generation, the resulting males are backcrossed to A. arabiensis wild-type females. In the bottleneck generation F3 hybrid males harbor an A. arabiensis X with a fraction (∼25% on average) of autosomal regions expected to be homozygous for the arabiensis background (red arrow). (B) Observed number of eggs and progeny arising for each indicated cross where the F1 hybrid females had either an A. gambiae mother (top) or an A. arabiensis mother (bottom). The asterisk indicates strain GY1 or GY2, respectively. In the bottleneck generation (F3), males were crossed to either pure species arabiensis females (♀A), or crossed to female hybrids with decreasing levels of A. gambiae genome content (♀H(F2), ♀H(F3)).