Fig. 3.
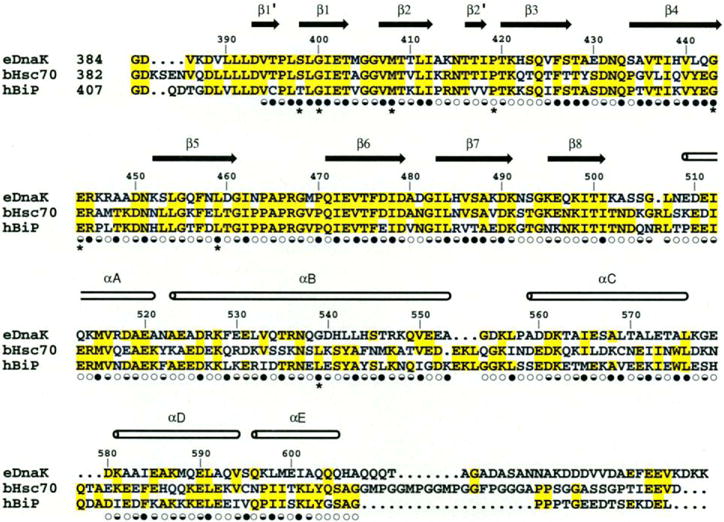
Structure-based sequence alignment of the C-terminal substrate binding unit of E. coli DnaK, bovine hsc70, and hamster BiP. The secondary structure assignments, based on main-chain hydrogen bonding pattern, are shown as arrows and cylinders. Solvent accessibility is indicated for each residue by an open circle if the fractional solvent accessibility is greater than 0.4, a half-closed circle if 0.1 to 0.4, and a closed circle if less than 0.1 (30). Residues changed in binding-defective genetic mutations are identified by asterisks (51).
