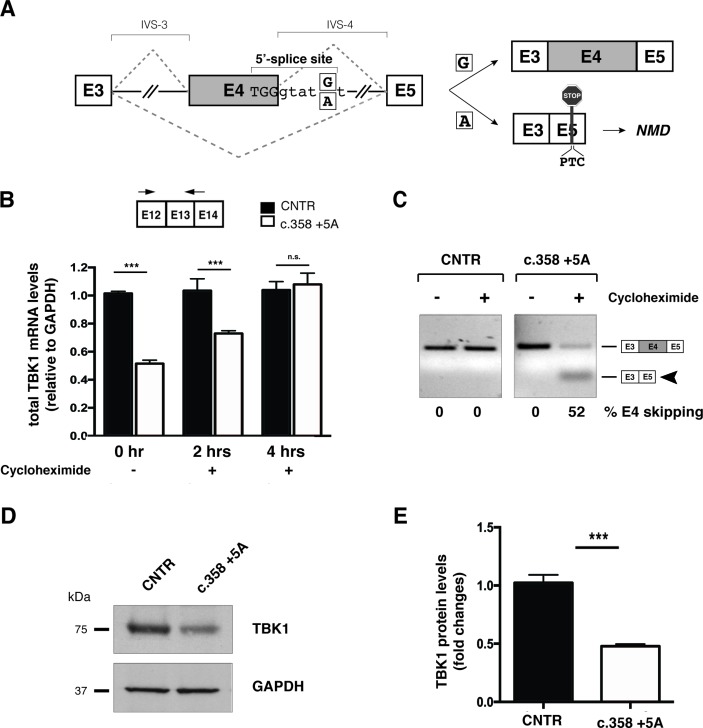
Figure 1.
c.358+5A variant causes aberrant splicing of exon 4 leading to TBK1 haploinsufficiency. (A) Schematic representation of wild-type (c.358+5G) and mutated (c.358+5A) sequences. Exons are depicted as boxes (E3, E4 and E5) and introns (interVening sequence (IVS-3 and IVS-4) as lines. The sequence surrounding exon 4 donor splice site of the wild-type G and mutant A is indicated (position +5). The two possible splicing patterns (E4 inclusion or skipping) are indicated with diagonal dashed lines and the two possible resulting splicing products schematically reported on the right of the panel. (B) Analysis of total TBK1 mRNA levels by quantitative RT-PCR. Total RNA was extracted from primary control and c.358+5G/A fibroblasts at different time points (0, 2 and 4 hours) after cycloheximide administration. Arrows correspond to TBK1_E12/13 Fw and TBK1_E13 Rev quantitative PCR; primers. Results are expressed as mean±SD of three different experiments performed in triplicate. (C) TBK1 exon 4 splicing assay. Total RNA from samples in (B) was analysed by semiquantitative RT-PCR using TBK1_E2/3 Fw and TBK1_E5 Rev to detect inclusion or skipping (arrowhead) of exon 4 (see schematic diagrams in A). Band intensities were quantified and the percentage of exon 4 skipping is reported under each lane of the representative gel. (D) Immunoblot analysis of TBK1 protein levels in control and c.358+5G/A patient primary fibroblasts. GAPDH serves as a loading control. (E) Quantification of band intensity in (D) shows fold change reduction in c.358+5G/A fibroblasts relative to controls (mean± SD from three independent experiments). ***p-value <0.001. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; NMD, non-sense mediated decay; n.s., not significant (Student’s t-test); RT-PCR, real-time PCR; TBK1, TANK-binding kinase 1, CNTR: Control.