Figure 2.
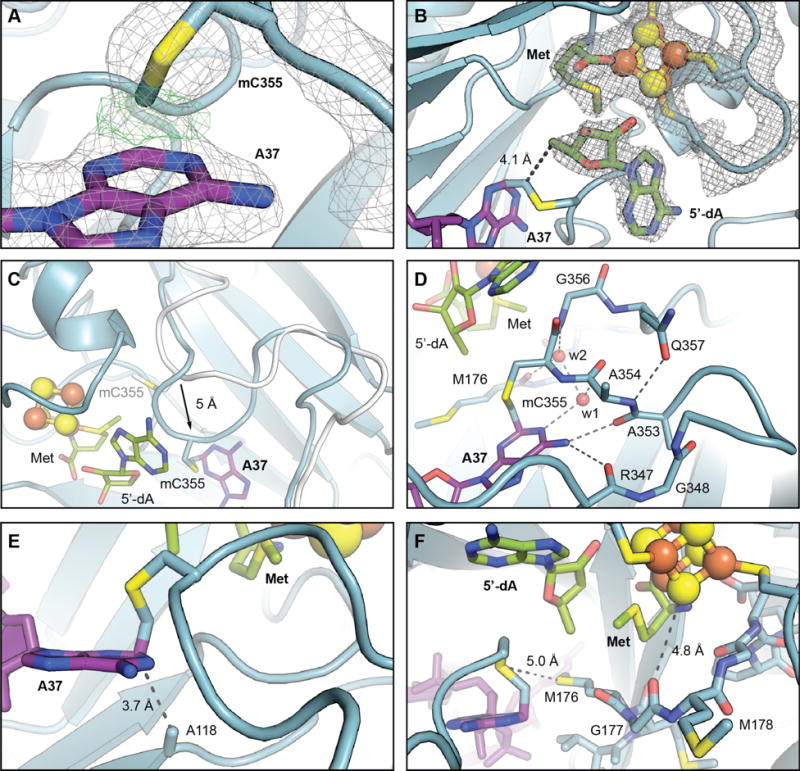
Views of the active site in the structure of the RlmN C118A-tRNAGlu in vitro cross-link. (A) A zoomed-in view of mC355 and A37 with selected residues and cofactors shown in stick format and colored by atom type. A 2Fo-Fc electron density map (gray mesh, contoured at 2.0 σ) and an omit map (green mesh, contoured at 4.0 σ) for the covalent bond between A37 C2 and the mCys355 Cδ are shown in overlay. (B) 2Fo-Fc electron density map for RlmN residues 125–132 (CX3CX2C motif, which binds [4Fe-4S] cluster), the [4Fe-4S] cluster, methionine, and 5′dA. The distance between the 5′-carbon of 5′-dA to the mC355 Cδ, consistent with H• abstraction via a 5′-dA• necessary to initiate cross-link formation, is indicated by a dashed line. (C) Overlay of loop from the wt RlmN x-ray structure with SAM (white, PDB accession code 3RFA) with the RlmN C118A in vitro cross-link (blue), illustrating the 5Å shift in position of the loop backbone upon interaction with the tRNA substrate. Selected amino acid side chains, nucleic acid bases, 5′-dA, and methionine are shown in stick format. The [4Fe-4S] cluster is illustrated as a ball-and-stick model. (D) A detailed view of the interactions involved in loop repositioning. Hydrogen bonding interactions are illustrated with dashed lines and ordered water molecules are shown as red spheres. (E) A zoomed-in view of the loop containing mC355 with the covalent cross-link to C2 of A37 shown in sticks. The distance between C2 of A37 and A118, consistent with assignment of C118 as a proton acceptor in the wt RlmN reaction, is indicated by a dashed line. (F) An alternate view of the active site with the conserved MGMGE motif and other selected amino acids displayed in stick format. Interactions potentially important in thiyl radical stabilization or in electron transfer mediated by the [4Fe-4S]1+ are shown as dashed lines.
