Figure 1.
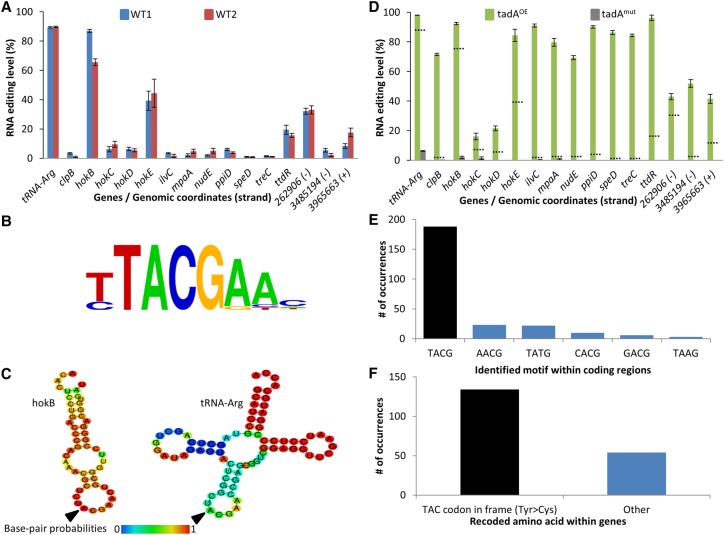
RNA editing occurs in E. coli and it is mediated by tadA. (A) RNA-seq (and DNA-seq) from two WT E. coli strains (Top10 and MG1655-EcM2.1, blue and red, respectively) reveals 15 novel A-to-G(I) RNA editing sites in E. coli in addition to the known editing site in tRNA-Arg. Notably, all sites found in known genes (12 out of the 15 sites) recode a tyrosine (TAC) into a cysteine (TGC) codon. The three RNA editing sites that do not occur in known ORFs are denoted by their genomic coordinates and genomic strand (+ or −). RNA editing levels are defined here as the number of reads with a G at the position out of all reads that cover the position. RNA samples were extracted in mid-log phase at OD600 ∼ 0.7. (B) All sites share a common four-base DNA motif which is identical to tadA's recognition motif. (C) RNA secondary structure modeling predicts that edited sites are embedded within a loop. Here, the secondary structure of hokB (as well as tRNA-Arg) is presented (the RNA secondary structure modeling of all other targets found in this work is shown in Supplemental Fig. S2). (D) Overexpressing (green) or mutating (gray) tadA increases or reduces the editing level, respectively. Dotted lines represent the average editing levels measured for each gene in the two WT strains. RNA samples were extracted in mid-log phase at OD600 ∼ 0.5–0.6. (E) Overexpression of tadA reveals additional putative editing sites, of which 75% are embedded within the canonical motif (TACG, black bar), while the rest deviate by one base from the canonical motif. (F) Out of 188 editing sites which occur within genes, 134 (black bar) recode a Tyr into a Cys codon (71%). Error bars in parts A and D represent standard errors of measuring editing level in a given coverage. Exact values can be found in Supplemental Tables S1 and S2.