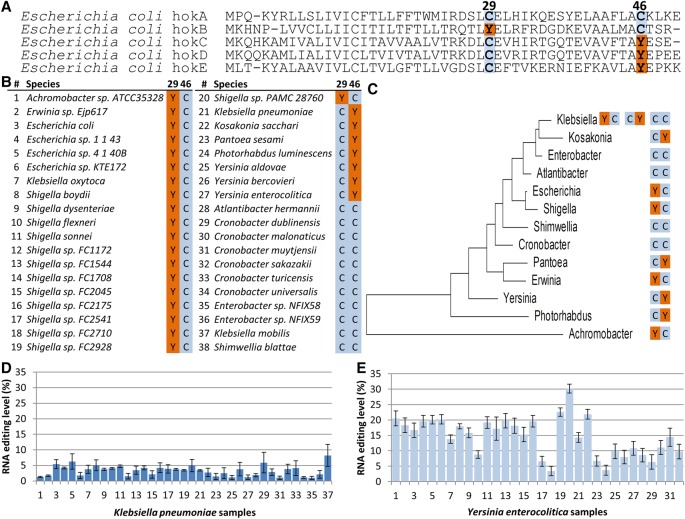
Figure 2.
Evolutionary analyses suggest an interplay between the recoded and a hard-coded cysteines in hokB. (A) Multiple sequence alignment of five hok proteins encoded by the E. coli genome (NC_000913.3). The hokB edited version recapitulates the cysteine at position 29 which is hard-coded in the genome of all other hok protein family members. Symmetrically, hokC, hokD, and hokE editing sites (position 46) recapitulate the cysteine at the same position of hokB. (B) Multiple sequence alignment of hokB of a representative nonredundant set of orthologs from bacterial species harboring an annotated hokB gene suggests interplay between peptide residues at positions 29 and 46. Notably, all the Tyr codons at position 29 or 46 are encoded by the editable codon (TAC, embedded within the TACG motif). The complete alignment can be found in Supplemental Table S4. (C) A maximum likelihood phylogenetic tree based on the 16S rRNA gene, showing the amino acid composition at hokB's positions 29 and 46 in each bacterial genus with species harboring an annotated hokB. (D,E) RNA editing in hokB was identified in publicly available Klebsiella pneumoniae (37) and Yersinia enterocolitica (32) samples with sufficient coverage (≥51×) of RNA reads and at least two reads supporting an editing event. This editing event is predicted to recode position 46 (Tyr>Cys) in hokB. SRA accession numbers can be found in Supplemental Tables S5 and S6. Error bars represent standard errors of measuring editing level in a given coverage.