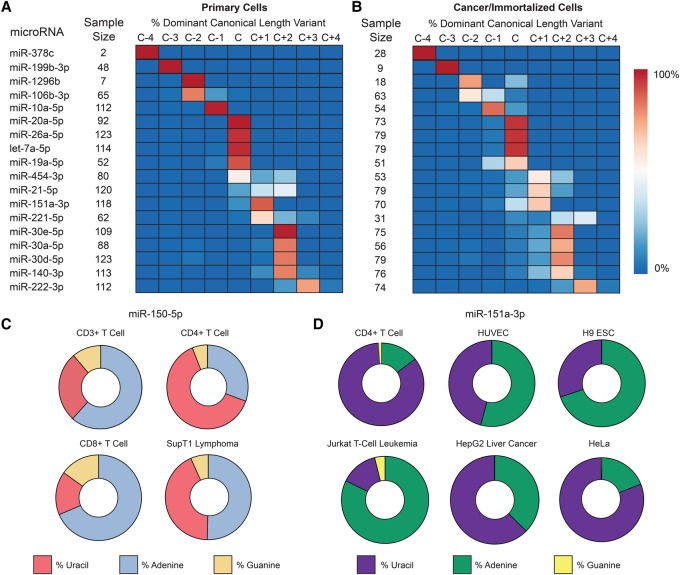
Figure 4.
IsomiRs are a challenge to characterizing microRNA levels. (A) Among primary cells, the most abundant (dominant) sequence for many microRNAs differs in length from the canonical “C” miRBase.org v21 sequence by up to 4 bases (C−4 to C+4). Between cell types, length diversity is also present, as evidenced by microRNAs that are not entirely of one length. Eighteen representative microRNAs from 556 in total. (B) The general features of microRNA length among cancer/immortalized cells are similar, but the microRNA processing in these cells skews toward randomness. See also Supplemental Figure 12. (C) miR-150-5p, a lymphocyte-specific microRNA, shows a diversity of nontemplated nucleotide addition at the +1 site on the 3′ end. Cytosine is the templated (genomic) nucleotide at this position and is not shown. (D) miR-151a-3p, a ubiquitous microRNA, has marked variation in the first nontemplated nucleotide addition. Cytosine is again the templated nucleotide at this position.