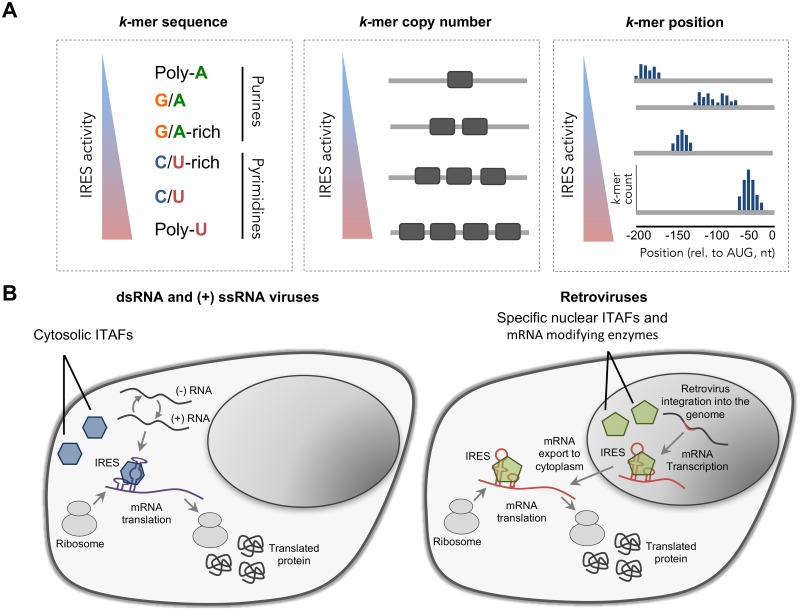
Fig 6. Summary of the sequence features associated with IRES activity.
(A) Illustration of the sequence features found by our models and their association with IRES activity: (left) k-mer sequence, (middle) the number of sites of a k-mer, and (right) the position of the k-mer relative to the AUG start codon. (B) Illustration of the different life cycles of (left) dsRNA/(+) ssRNA viruses and (right) Retroviruses which may have led to differences in their IRESs sequence features. Retroviruses are integrated into the host genome and RNA-PolII transcribes their mRNA in the nucleus. Thus, their IRES elements are exposed to the nuclear environment including mRNA modifying enzymes (methylation, pseudouridylation etc) and nuclear specific ITAFs that can shuttle with the mRNA to the cytoplasm to facilitate cap-independent recruitment of the ribosome. In contrast, dsRNA and (+) ssRNA viruses that spend their entire replication cycle in the cytoplasm are exposed to cytosolic factors, which in turn can facilitate cap-independent recruitment of the ribosome.