Figure 2.
Identification of PATL2 Variants in Individuals with a Phenotype Involving Oocyte Maturation Arrest
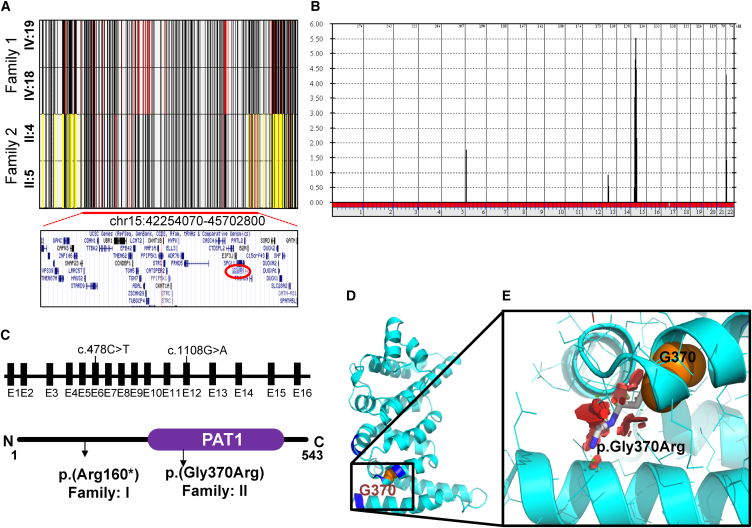
(A) Shared haplotypes (chr15: 42,254,070–45,702,800; UCSC Genome Browser hg19) of the affected members from family 1 (IV:19 and IV:18) and family 2 (II:4 and II:5). Yellow lines indicate heterozygous SNPs. Red lines indicate rare homozygous SNPs. The lower panel shows genes that are within the shared ROH region. The position of PATL2 (chr15: 44,966,961–45,003,514; UCSC Genome Browser hg19) is highlighted with a red circle.
(B) Genome-wide linkage analysis showing a chr15 peak with a pLOD score of 5.5 (16 individuals are included; see Figure S2 for details).
(C) Genomic context and structural presentation of PTAL2, including the two variants.
(D and E) The PAT1 domain contains the residues (Arg301, Leu302, Arg368, Ala327, Arg402, and Arg403) corresponding to the PATL1 residues involved in RNA binding (shown in dark blue). (D) Based on the structure of the corresponding region of PATL1 (PDB: 2XEQ; 34% sequence identity), the secondary-structure representation of the homology model of the C-terminal domain of PATL2 (residues 299–540) is shown. Gly370 is shown as orange spheres. (E) Magnified view of the outlined region in (D). In addition to Gly370 (orange), the putative p.Gly370Arg rotamer side chain (gray) with the least possible clashes (red discs) is shown, illustrating that p.Gly370Arg compromises the folding and function of PATL2.