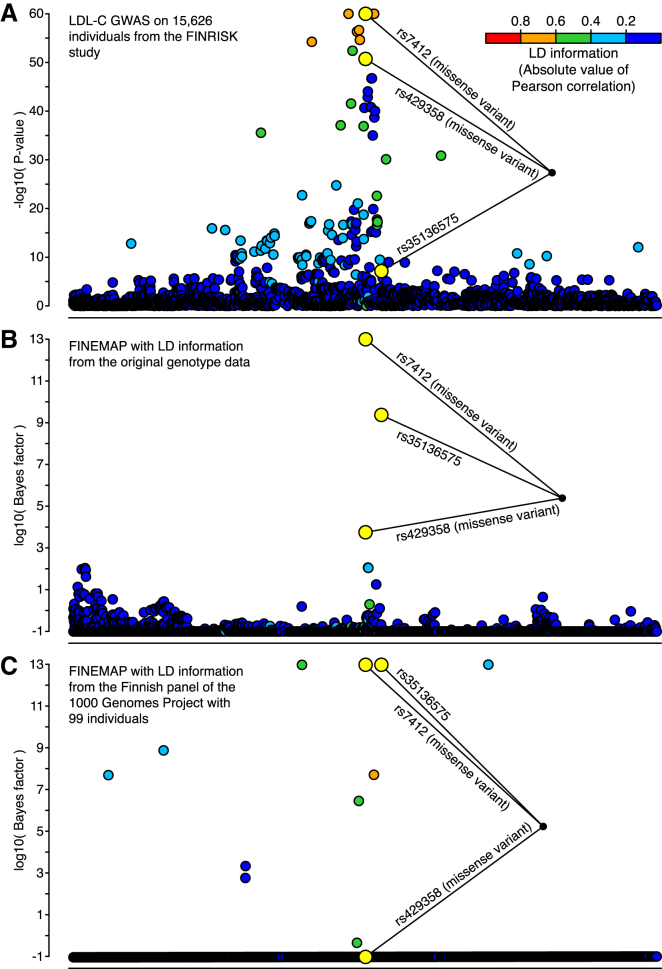
Figure 2.
Fine-Mapping the APOE Region Associated with LDL-C
Results are shown for 3,078 variants with a MAF above 1% and covering 1 Mb of the genome. Variants identified by a standard conditional analysis are highlighted in yellow. All other variants are colored with respect to their LD (absolute value of Pearson correlation) with the lead variant rs7412.
(A) Negative log10 p values for each variant from a LDL-C GWAS on 15,626 individuals from the FINRISK study.
(B) Bayes factor (log10) for assessing the causality of each variant by a FINEMAP analysis using the summary statistics from the LDL-C GWAS and the LD information from the original genotype data.
(C) Bayes factor (log10) for assessing the causality of each variant by a FINEMAP analysis using the summary statistics from the LDL-C GWAS and the LD information from the reference genotypes of 99 Finns in the 1000GP.