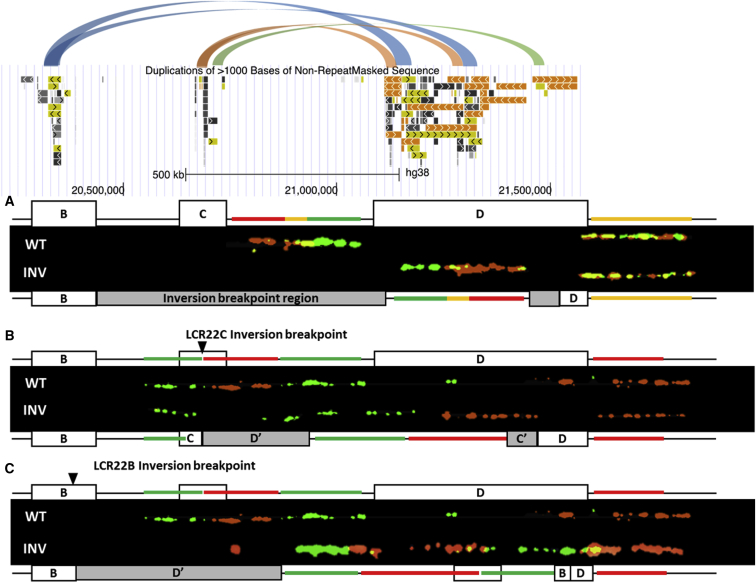
Figure 1.
Design and Fluorescence Patterns
(A) First probe design aligned with schematic representation of the LCR22B-D region. LCR22s are depicted with the Segmental Dups track10,33in UCSC.34 Locations of known segmental duplications between LCR22B, LCR22C, and LCR22D are shown as connecting, colored lines. Red and green oligos (hg19, chr22:21,104,802–21,311,933 and chr22:21,223,781–21,424,739) partially label the unique sequence proximally from LCR22D and have 50 Kb of overlapping sequence depicted by the central yellow bar. A reference probe distal to LCR22D (chr22:21,931,955–22,124,705) is labeled with a green-red mix, shown as a yellow bar. Oligos hybridized to a reference allele show the expected wild-type (WT) sequence of red, overlapping yellow, and green signal strings. Subsequently, an unlabeled gap marks the position of LCR22D, followed by the mixed reference signal. Inversion (INV) signals show a signal sequence of green, overlapping yellow, and red, followed by the LCR22D gap and the mixed reference probe.
(B) Second probe design aligned with the LCR22B-D region. An additional green probe, starting proximally from LCR22C to the middle of the repeat (chr22:20,499,302–20,699,303), is added. From this location, a red (chr22:20,699,530–20,899,531) and a green (chr22:20,890,041–21,103,073) probe are designed to consecutively label 200 Kb each. A red reference probe marks the region distal to LCR22D (chr22:21,572,091–21,773,001). Probe patterns suggest a LCR22C–D inversion polymorphism.
(C) A probe pattern suggesting a LCR22B–D inversion.