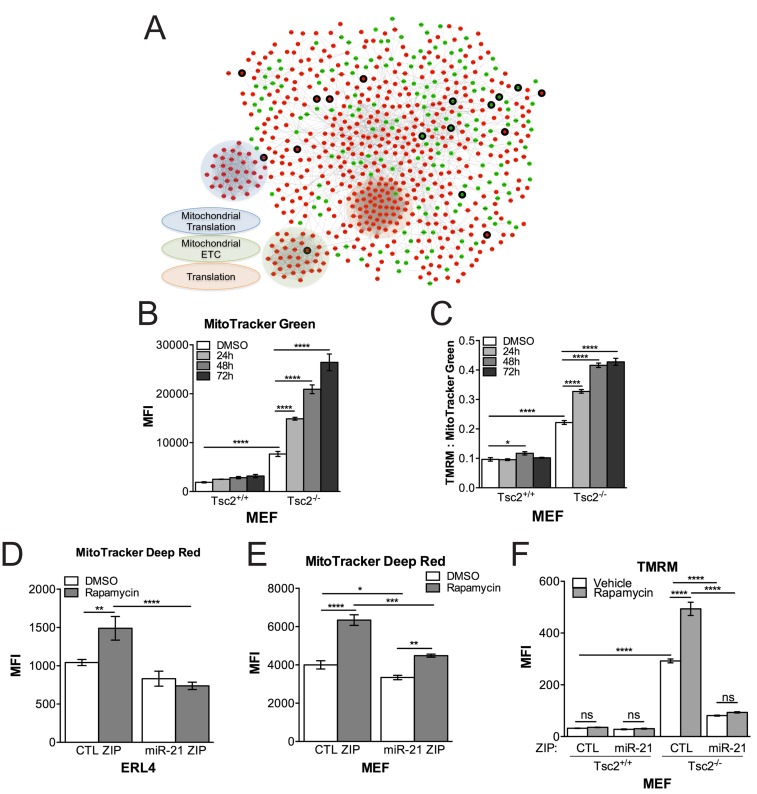
Figure 3. Network-based RNAseq analysis implicates miR-21 in mitochondrial processes.
A., Network analysis using the consolidated interactome of differentially expressed genes between miR-21 ZIP and CTL ZIP MEFs treated with rapamcyin (20nM, 24h). red upregulated, green-downregulated, thick border- predicted targets of miR-21. B., Mitochondrial content was increased in Tsc2-/- MEFs treated with rapamycin (20 nM) in a time dependent manner in contrast to Tsc2+/+ MEFs. C., Rapamycin increases mitochondrial polarization in Tsc2-/- MEFs as assessed by TMRM normalized to MitoTracker Green. D., E., Cells stably expressing a miR-21 inhibitor have lower mitochondria content by MitoTracker Deep Red particularly following rapamycin in ERL4 and MEF cells.F, miR-21 inhibition reduces mitochondrial membrane polarization. Data presented as mean+/- standard deviation. Statistical significance was assessed by Two-Way and One-Way ANOVAs with Bonferroni correction with *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.