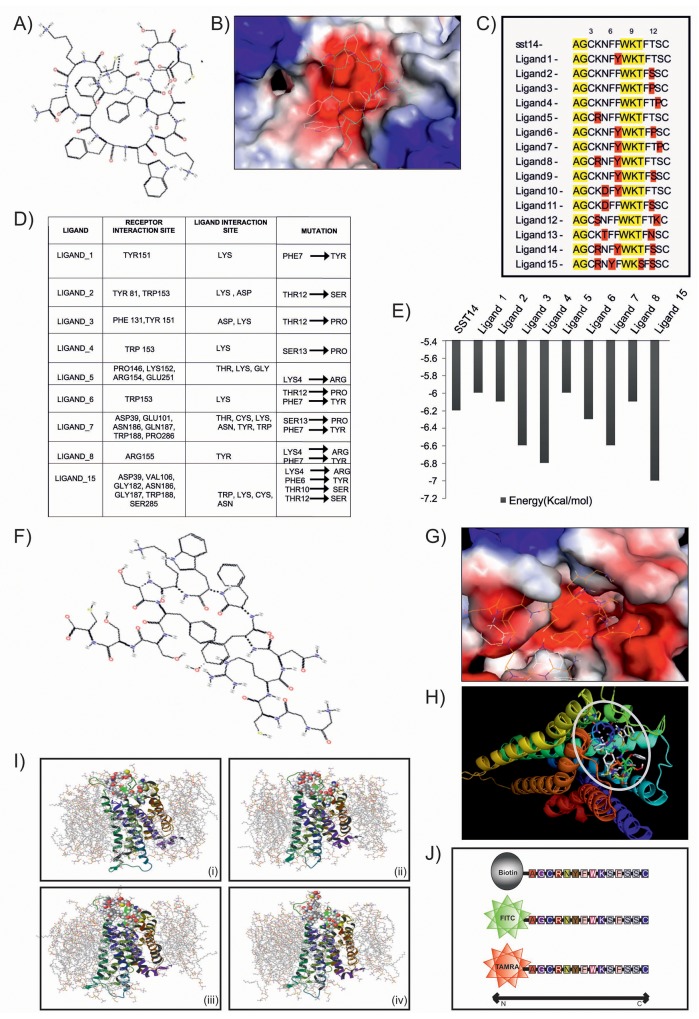
Figure 1. Design and in silico validation of potent peptide ligand for targeted delivery.
(A) & (B) Image represents chemical structure of SST14 and its binding with SSTR2. (C) & (D) Figure and table show mutated peptides and interaction of them with SSTR2. (E) Graph illustrates comparison of binding energy of various peptides with SST14. (F) & (G) Images represent chemical structure of ligand_15 and its efficient binding with SSTR2. (H) Patch dock result shows docking of ligand_15 (stick model) on SSTR2 (ribbon model). (I) Image represents molecular dynamic simulation of SSTR2- Ligand_15 complex at (i) 0 ns, (ii) 1.5 ns, (iii) 3.1 ns, and (iv) 4.8 ns. (J) Schematic diagram shows different types of tagging (biotin, FITC, TAMRA) at the N terminus region of synthesized SSTR2 peptide.