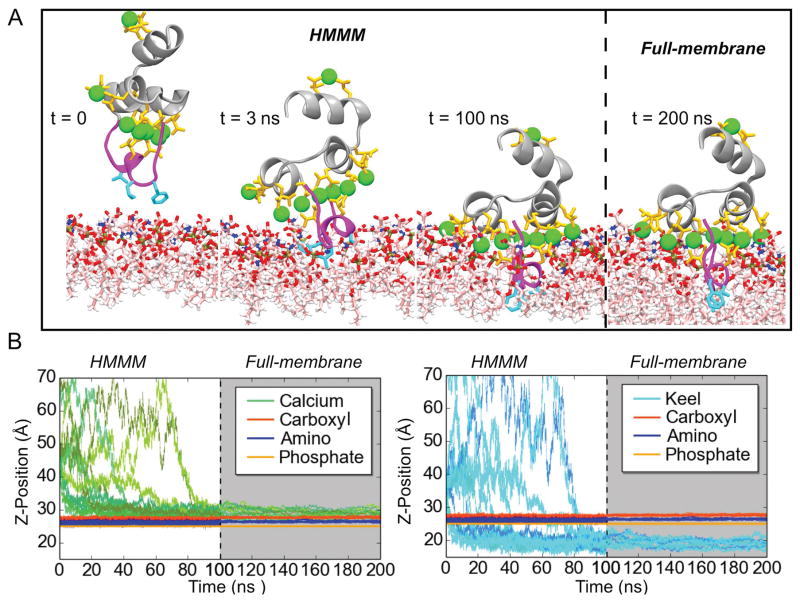
Figure 3.
Membrane binding and insertion of the FX-GLA domain. A) Binding of the FX-GLA domain at different time points along a representative trajectory. Until 100 ns (indicated by the dashed line), the domain is simulated in an HMMM membrane representation. Then, the membrane is converted to a full membrane. B) Relative positions on the z-axis of the COM of lipid phosphate, carboxyl and amino groups (both panels) and GLA domain residues (Ca2+ ions in left panel; backbone atoms of keel residues (PHE4, LEU5, and VAL8) in right panel). See Methods for atoms included in the phosphate, carboxyl, and amino groups. Grey background indicates portions of the trajectories with a full membrane. Data in this figure refer only to the 14 properly bound FX-GLA domain. Refer to Figure S3 for information on the 13 trajectories in which proper binding was not achieved within the time scale of the HMMM simulations (100 ns). See Methods for the definition of “proper” membrane binding.