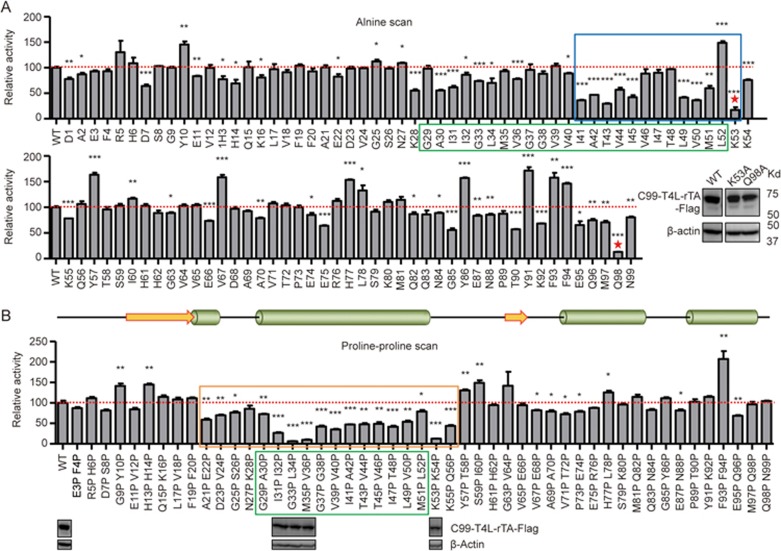
Figure 3.
Validation of minimum substrate. (A) Scanning alanine mutagenesis experiments. The TM domain and a C-terminal portion of the TM domain plus the TM boundary residue (amino acids 41-53) are highlighted by green and blue boxes, respectively; K53 and Q98 are marked by red stars. The relative protein expression levels of C99 WT, K53A and Q98A are shown by western blot in the inlet panel. (B) Scanning double proline mutagenesis experiments. The minimum region (E22-K55) required for efficient γ-secretase cleavage was marked by an orange box. Top: Predicted secondary structure (predicted by PSIPRED Protein Sequence Analysis)40. The relative protein expression levels of C99 WT and some crucial mutations are shown at the corresponding positions. Error bars=SEM, n=3, P-values (two-tailed Student's t-test versus WT): *P<0.05; **P<0.01, ***P<0.001.