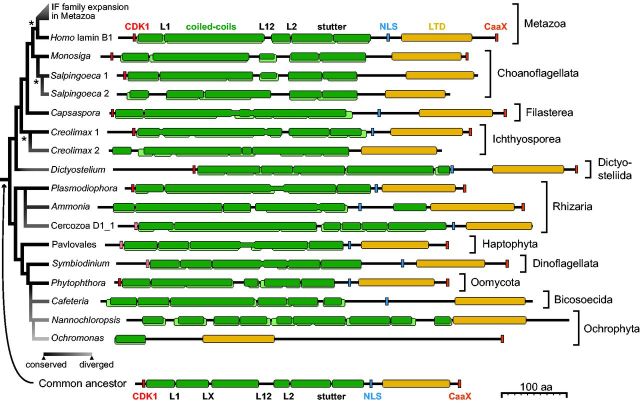
Fig. 2.—
Comparison of lamin architecture across eukaryotes. Sequence motifs and structural elements are highlighted: CDK1 phosphorylation consensus sequence in red, classical NLS in blue, CaaX motif in orange, and Ig-like LTD domain highlighted in yellow. Coiled-coil regions and the interruptions in the heptad repeats were predicted in Marcoil (dark green) and Pcoils (light green). The heptad-repeat interruptions are named according to animal IF proteins. An additional interruption between L1 and L12 that was predicted in many of the lamins and was presumably present in LECA is marked as LX. See supplementary figure S3, Supplementary Material online, for sequence alignment and predicted pattern of heptad repeats in the filament domain and secondary structure elements in the LTD domain. The phylogenetic relationships between lamins are displayed on left, together with the level of divergence in respect to sequence, domain architecture and a presence of sequence motifs (grey scale). The asterisks point to the lineage-specific duplications of lamin gene.