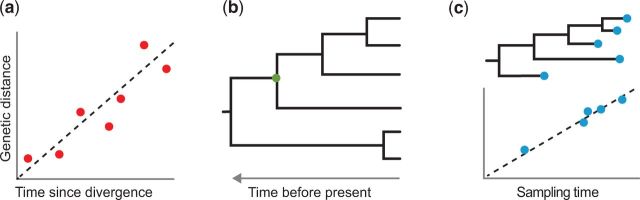
Fig. 1.—
Methods used to estimate evolutionary timescales from genomic data. (a) Linear regression of pairwise genetic distances against time since divergence. Each data point in this plot represents a pair of taxa, with their divergence time inferred from the fossil record or from the age of a geological event that is presumed to be associated with the evolutionary divergence. Fitting a line through these points involves the assumption that genetic change accumulates at a constant rate through time, with the slope presenting an estimate of this rate. The line of best fit can be used to infer the timing of evolutionary divergence events, provided that a measure of genetic distance is available for the taxa in question. Molecular clocks based on linear regression have a number of weaknesses, including nonindependence of the data points and sensitivity to rate variation across lineages. (b) Phylogenetic analysis using a clock model. The tree is a chronogram with branch lengths measured in units of time. These methods usually involve models that explicitly describe the evolution of characters along the branches of the tree. Phylogenetic molecular clocks are calibrated by constraining the age of one or more nodes in the tree, such as the node indicated with a green circle, allowing the remaining node times to be inferred from the genetic data. (c) Root-to-tip distances computed from a phylogram, plotted against the ages of the sequences. A regression line is fitted through these data points, with the slope of the line giving an estimate of the evolutionary rate. This method is often used in analyses of time-structured sequence data, such as those from rapidly evolving viruses.