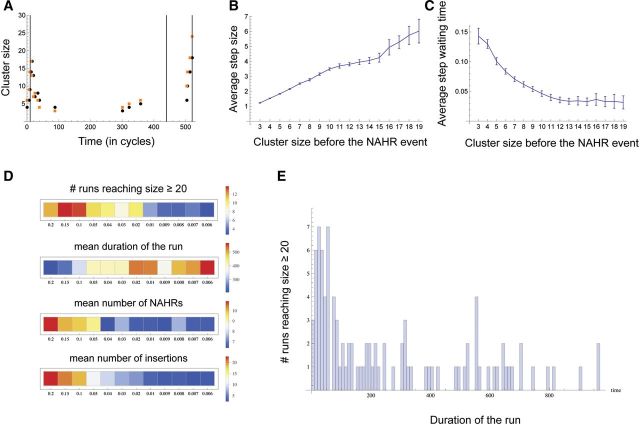
Fig. 6.—
Computer simulation of gene family expansion. (A) An example of a single simulation run that was terminated upon the cluster reaching size = 20. Parameter values were: ι = 0.14, µ = 0.4 and u = 0.02. Black and orange dots represent the cluster sizes immediately before and after NAHR, respectively. Timings of the fresh LINE insertions are shown by the vertical lines. (B) The average size of gain or loss (i.e., step) in the gene cluster as a result of NAHR for each cluster size category (i.e., immediately before the NAHR occurred). The initial cluster sizes are shown on the X axis and 95% confidence intervals for the means are indicated by the vertical error bars. (C) The average run duration for each cluster size category. The absolute values of run duration depend on the parameter values chosen, whereas, here, we are interested in relative comparison of durations among the cluster size categories. To remove the effects of the parameters, for each combination of the parameter values, the run durations were normalized so that they sum to 1. The data in (B) and (C) were pooled over 48,000 runs, with µ varying from 0.1 to 0.4 and u varying from 0.006 to 0.2. (D) The effect of the rate of insertion of fresh LINEs, u, on (1) the number of “successful” runs that reached size ≥20, shown as the proportion of the 1,000 runs, (2) the average duration of the successful runs, (3) the mean number of NAHR in the successful runs and (4) the number of fresh LINE insertions (only the successful runs were considered). (E) An example of the distribution of the times of the last NAHR event among the successful runs. Parameter values were the same as in (A). Note the outlier runs where the last NAHR occurred much later then average.