Fig. 1.
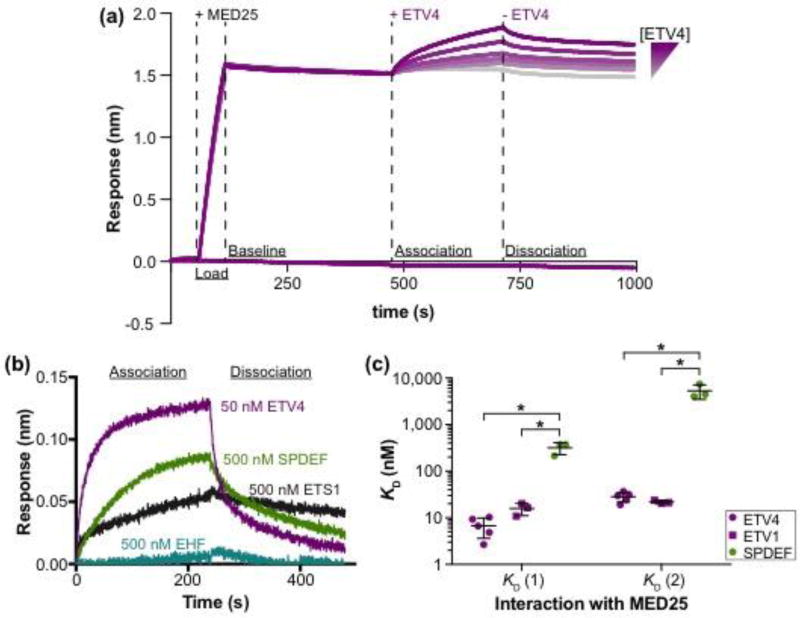
High-affinity interaction with MED25 is specific to the ETV1/4/5 subfamily of ETS factors. (a) Representative ETV4-MED25 sensorgrams using biolayer interferometry. Streptavidin sensors were loaded with the biotinylated ACID domain of MED25391–553 (+ MED25), followed by the addition (+ ETV4), then removal (− ETV4) of six different concentrations of ETV4 in order to measure the association and dissociation, respectively, between ETV4 and MED25 ACID. The bottom purple line is a control sensor with no MED25 loaded and the highest concentration of ETV4 added during the association step. (b) Representative sensorgrams of binding assays with a single concentration of ETV4 (50 nM), SPDEF (500 nM), ETS1 (500 nM), and EHF (500 nM). Only the association and dissociation phases of the sensorgram are shown. ERG (500 nM) and ETV6 (500 nM) were also tested but showed no detectable association; each ETS factor was tested twice. (c) Equilibrium dissociation constants (KD) for the interaction between MED25 ACID and SPDEF, ETV1, and ETV4. Two KD values, denoted KD (1) and KD (2) with KD (1) being the higher-affinity interaction, are reported as these interactions better fit a 2:1 (ETS:MED25) binding model (Fig. S1c). KD values were determined using a group fit with six different concentrations of each ETS factor (Fig.1a and Fig. S1a,b). Circles and squares represent the KD determined from a single, six-concentration experiment. Horizontal lines represent the mean and standard deviation for three to five replicate experiments. KD, ka, and kd values for these interactions are summarized in Table S1. “*” Indicates p < 0.05 in a Mann-Whitney U test.