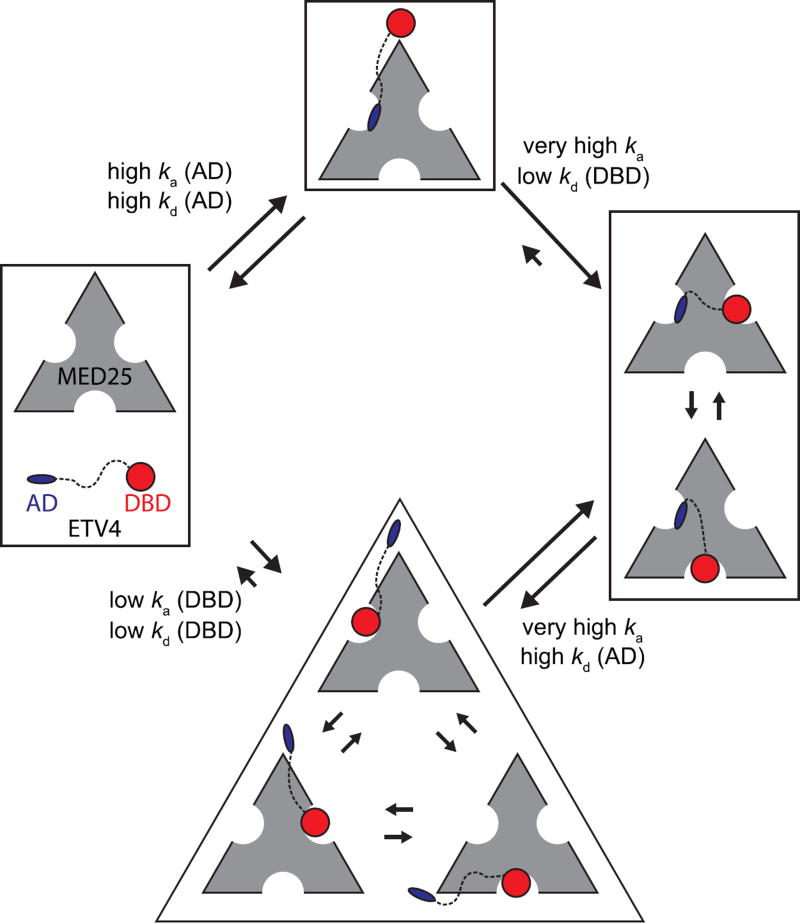
Fig. 10.
Model for the multivalent ETV4-MED25 interaction. In the predominant pathway (top) the first step of interaction involves the activation domain (AD) binding to MED25 Site 1 due to its drastically faster association compared to the interaction between MED25 and the DNA-binding domain (DBD, bottom). The association and dissociation rate constants, ka (AD), kd (AD), ka (DBD), and kd (DBD) for the first binding steps are assumed equivalent to those measured for the isolated AD-MED25 and DBD-MED25 interactions, respectively. In the second step of the predominant pathway, the DBD of the bound ETV4 can only bind to the two MED25 sites that are not occupied by the AD (right). The ETV4-MED25 interaction would then be in equilibrium between two states with the DBD alternately occupying Sites 2 and 3 on MED25. The predominant pathway for dissociation of ETV4-MED25 will be limited by the slower release of the DBD. ETV4 DBD binding to MED25 is not predicted to be the predominant first step for the interaction for full-length ETV4 and MED25 (bottom), due to the drastically slower association rate (bottom). However, in circumstances where the AD is bound to another protein, or in oncogenic translocations of ETV4 that do not have the N-terminal AD, binding of the DBD to MED25 would still be a viable mode of interaction. The size of the arrows indicating association and dissociation between the unbound and AD-MED25 or DBD-MED25 complexes, and dissociation between the ETV4-MED25 and AD-MED25 or DBD-MED25 complexes, is representative of the measured rate constants for these steps. Other arrows are qualitatively sized based on mutational data and interpretations, and should not be considered as measured values.