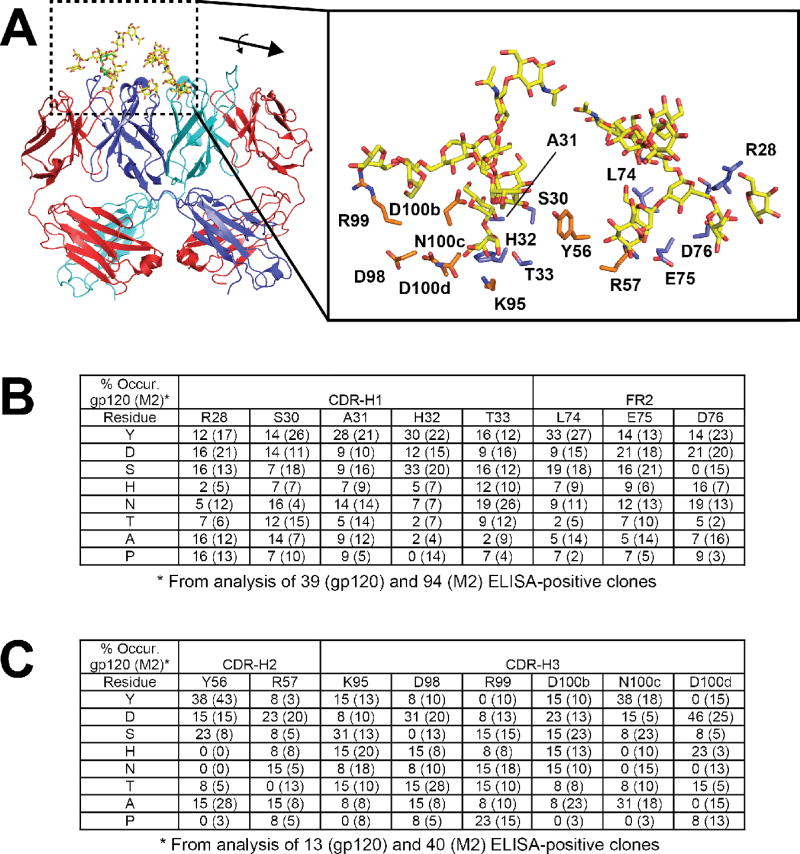
Fig. 4. Limited diversity 2G12 library design and results from selection.
(A) Positions that were targeted for mutagenesis involve direct contact residues for glycans at both the primary and secondary sites; blue side chains in the close-up were for Lib1 and orange side chains were for Lib 2. All of the residues were allowed to vary among eight residues (Y/D/S/H/N/T/A/P) using the NMC degenerate codon. For clarity, only one antigen for each is shown. (B and C) Percent occurrence of the eight possible residues following selection against gp120 at each position for Lib1 (B) and Lib2 (C). For reference and to control for expression biases, a parallel selection was performed against M2, an anti-FLAG antibody, and the percent occurrence listed in brackets.