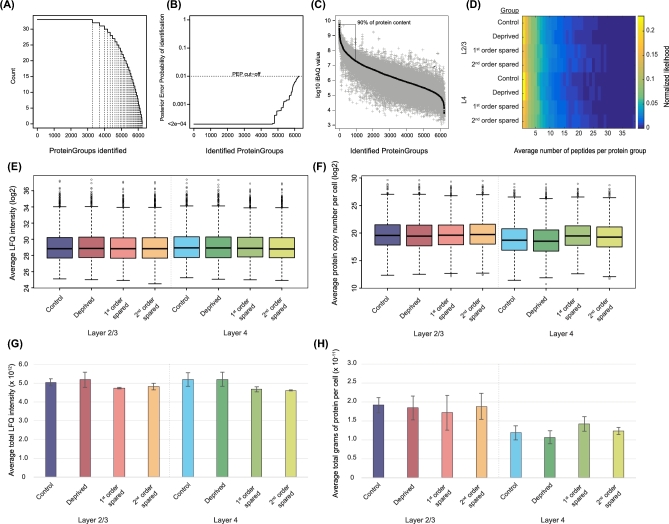
Figure 3:
Quantification of protein groups across all samples. Control/deprived, C column; first-order spared, B/D columns; second-order spared, A/E columns (see Fig. 1). (A) Number of observations per protein group in the entire dataset. (B) Confidence of protein group identification across samples. (C) Protein content versus identified protein groups. For every protein group, all measured iBAQ values are plotted in grey, with the median value in black. (D) Averages and variances of peptides per protein group in each experimental group. (E) Box plot of LFQ intensity averages across samples within each group. (F) Box plot of protein copy numbers per cell (inferred as in [6]) averaged across samples within experimental groups. (G) Summed LFQ intensities averaged within experimental groups. (H) Total mass of identified proteins per cell, averaged within experimental groups. The inferred protein copy number per cell was divided by Avogrado's number (6.0221409 × 1023) and then multiplied by the protein mass in kilodaltons (kDa), yielding the total mass of identified proteins per cell.