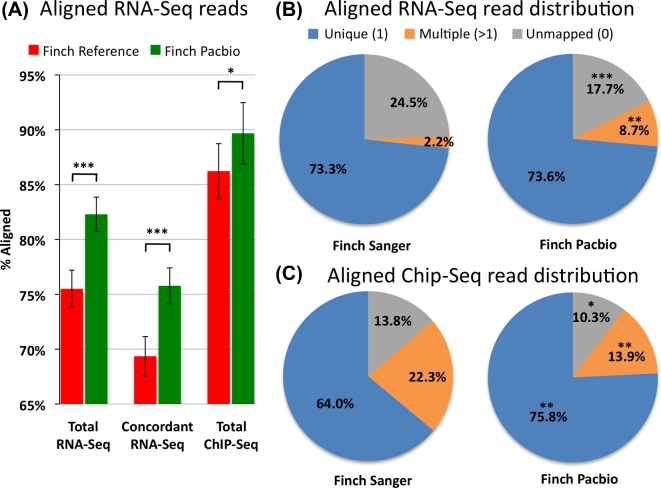
Figure 2:
Transcriptome and regulome representation within assemblies. (A) Percentage of RNA-Seq and H3K27Ac ChIP-Seq reads from the zebra finch RA song nucleus mapped back to the zebra finch Sanger-based and PacBio-based genome assemblies. (B) Pie charts of the distributions of the RNA-Seq reads mapped to the zebra finch genome assemblies. (C) Pie charts of the distribution of ChIP-Seq reads mapped to the zebra finch genome assemblies. *P < 0.05; **P < 0.002; ***P < 0.0001; paired t test within animals between assemblies; n = 5 RNA-Seq and n = 3 ChIP-Seq independent replicates from different animals.