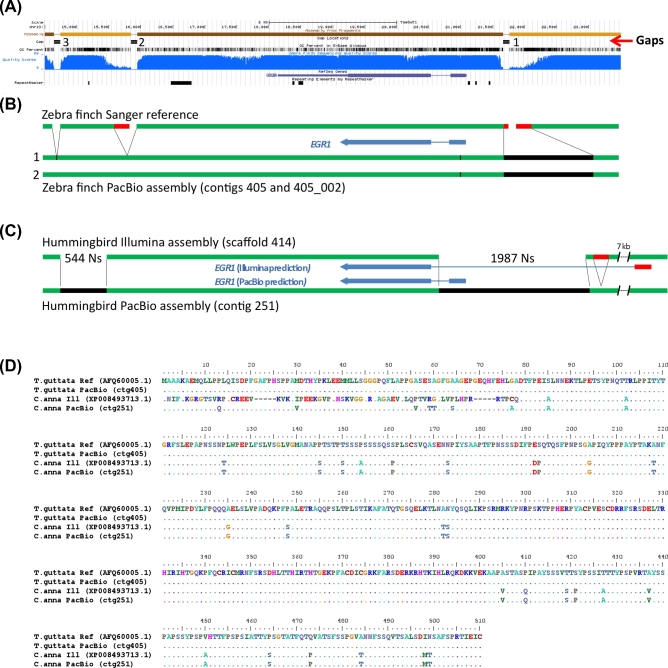
Figure 3:
Comparison of EGR1 assemblies. (A) UCSC genome browser view of the Sanger-based zebra finch EGR1 assembly, highlighting (from top to bottom) 4 contigs (light and dark brown) with 3 gaps, GC percent, nucleotide quality score (blue), RefSeq gene prediction (purple), and areas of repeat sequences. (B) Summary comparison of the Sanger-based and PacBio-based zebra finch assemblies, showing in the latter filling the gaps (black) and correcting erroneous reference sequences surrounding the gaps (red). Tick mark is a synonymous heterozygous SNP in the coding region between the primary (1) and secondary (2) haplotypes. Panels A and B are of the same scale. (C) Comparison of the hummingbird Illumina- and PacBio-based assemblies, showing similar corrections that further lead to a correction in the protein coding sequence prediction (blue). (D) Multiple sequence alignment of the EGR1 protein for the 4 assemblies (2 zebra finch and 2 hummingbird) in (B) and (C), showing corrections to the Illumina-based hummingbird protein prediction by the PacBio-based assembly.