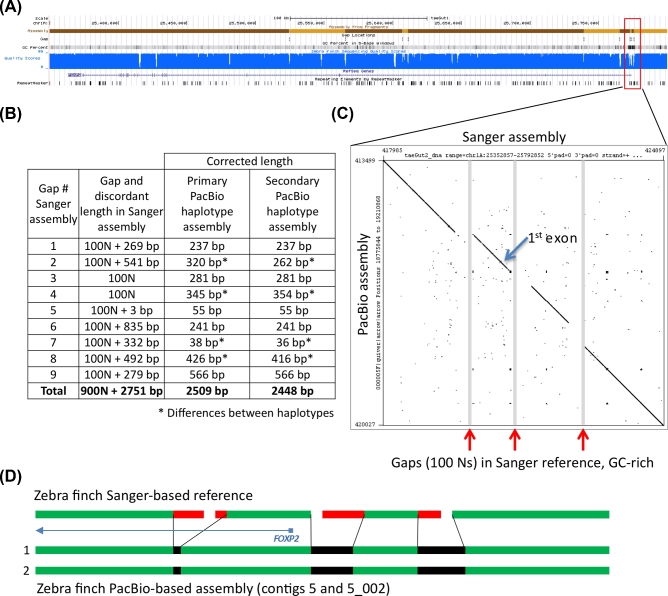
Figure 5:
Comparison of FOXP2 assemblies. (A) UCSC genome browser view of the Sanger-based zebra finch FOXP2 assembly, highlighting 10 contigs with nine gaps, GC percent, nucleotide quality score, RefSeq gene prediction, and repeat sequences. (B) Table showing the number of resolved and corrected erroneous base pairs in the gaps by the PacBio-based primary and secondary haplotype assemblies; the asterisk indicates differences between haplotypes. (C) Dot plot of the Sanger-based reference (x-axis) and the PacBio-based primary assembly (y-axis) corresponding to the 3 GC-rich region gaps immediately upstream and surrounding the first exon of the FOXP2 gene. (D) Schematic summary of corrections to the 3 gaps shown in (C) in the 2 haplotypes of the PacBio-based assembly. The protein coding sequence alignments are in Figure S13A.