Abstract
Background
Risk of prostate cancer-specific mortality (PCSM) is highly variable for men with adverse pathologic features at radical prostatectomy (RP); a majority will die of other causes. Accurately stratifying PCSM risk can improve therapy decisions.
Objective
Validate the 22 gene Decipher genomic classifier (GC) to predict PCSM in men with adverse pathologic features after RP.
Design, setting, and participants
Men with adverse pathologic features: pT3, pN1, positive margins, or Gleason score >7 who underwent RP in 1987–2010 at Johns Hopkins, Cleveland Clinic, Mayo Clinic, and Durham Veteran’s Affairs Hospital. We also analyzed subgroups at high risk (prostate-specific antigen > 20 ng/ml, RP Gleason score 8–10, or stage > pT3b), or very high risk of PCSM (biochemical recurrence in < 2 yr [BCR2], or men who developed metastasis after RP [MET]).
Outcome measurements and statistical analysis
Logistic regression evaluated the association of GC with PCSM within 10 yr of RP (PCSM10), adjusted for the Cancer of the Prostate Risk Assessment Postsurgical Score (CAPRA-S). GC performance was evaluated with area under the receiver operating characteristic curve (AUC) and decision curves.
Results and limitations
Five hundred and sixty-one men (112 with PCSM10), median follow-up 13.0 yr (patients without PCSM10). For high GC score (> 0.6) versus low-intermediate (≤ 0.6), the odds ratio for PCSM10 adjusted for CAPRA-S was 3.91 (95% confidence interval: 2.43–6.29), with AUC = 0.77, an increase of 0.04 compared with CAPRA-S. Subgroup odds ratio were 3.96, 3.06, and 1.95 for high risk, BCR2, or MET, respectively (all p < 0.05), with AUCs 0.64–0.72. GC stratified cumulative PCSM10 incidence from 2.8% to 30%. Combined use of case-control and cohort data is a potential limitation.
Conclusions
In a large cohort with the longest follow-up to date, Decipher GC demonstrated clinically important prediction of PCSM at 10 yr, independent of CAPRA-S, in men with adverse pathologic features, BCR2, or MET after RP.
Patient summary
Decipher genomic classifier may improve treatment decision-making for men with adverse or high risk pathology after radical prostatectomy.
Keywords: Radical prostatectomy, Adverse pathologic features, Prostate cancer-specific mortality, Genomic classifier, CAPRA-S
1. Introduction
For men with adverse pathologic features at radical prostatectomy (RP), there is considerable variability in risk of subsequent recurrence, metastasis and death, and time intervals between these progression events can be long [1]. This variability complicates decisions for adjuvant, salvage, and metastatic treatment. In recent years, there has been a rapid increase in the number of treatments for advanced prostate cancer (PCa), with growing interest in sequencing these treatments earlier in the disease course [2,3]. Because these agents are not without side effects, and because early development of resistance could preclude more effective use at a later stage, it is important to target novel and aggressive treatment regimens to men at highest risk of PCa-specific mortality (PCSM).
Currently, surgical pathology features are the primary means to identify men at highest risk of PCSM [4,5]. Despite the ability of these features to stratify PCSM risk, there remains considerable variability in outcomes when applied to individual men with PCa. To improve prognostic accuracy and clinical decision-making, several risk classifiers have been developed based on biomarker signatures alone or integrated with clinical features. One of the signatures most extensively tested and validated for predicting risk of metastasis in men at intermediate and high risk is the Decipher genomic classifier (GC), comprised of 22 gene expression markers derived from whole transcriptomic analysis of formalin fixed paraffin embedded tissue [6–9]. The GC generates a score from 0 to 1, with higher values associated with worse outcomes. Recently the GC was also shown to predict risk of PCSM in men with high risk based on preoperative prostate-specific antigen (PSA) levels or pathology [10]. Because that study was based on a relatively small number of men from a single institution, and with few PCSM events and short follow-up, we undertook a more extensive validation of the ability of the GC to predict PCSM.
2. Materials and methods
2.1. Patient cohort
The study population comprised four cohorts of PCa patients with adverse pathologic features, defined as RP Gleason score ≥7, RP stage pT3 or pN1, or positive surgical margins; per the study protocol patients with neoadjuvant therapy were excluded. The cohorts included 407 men from Mayo Clinic who underwent RP from 1987 to 2006, 355 from Johns Hopkins treated from 1992 to 2010, 179 from Cleveland Clinic treated from 1988 to 2008, and 113 from the Durham Veteran’s Administration Medical Center from 1991 to 2010, totaling 1054 with all required analysis variables, among whom there were 141 confirmed PCa deaths. Patients from the Mayo Clinic cohort did not include men used to train the original GC [6]. Patient follow-up after RP was not standardized among the four institutions, but differences were minor. The primary outcome was PCSM within 10 yr of RP (PCSM10); patients who died of PCa >10 yr after RP were considered censored (n = 29), and patients alive with less than 10 yr of follow-up (n = 493) were excluded. This resulted in a total of 561 patients with 112 PCSM10 (79% of all PCSM). Institutional review boards at the participating institutions approved the research protocol.
2.2. Specimen processing
Specimen selection and processing have been described previously [7,11–13]. Following microarray quality control using the Affymetrix Power Tools packages (Thermo Fisher Scientific Inc., MA, USA) [14], probeset summarization and normalization was performed utilizing the single channel array normalization algorithm [15]. Information about obtaining Decipher for routine clinical practice is in the Supplementary data.
2.3. Statistical analysis
The GC was calculated as a numeric value ranging from 0 to 1, based on each patient’s individual expression of the 22 genes integrated in a previously trained and validated signature [6]. The Cancer of the Prostate Risk Assessment Postsurgical (CAPRA-S) score was likewise calculated by applying PSA, RP Gleason score, and RP stage values to a previously validated algorithm, producing a score ranging from 0 to 12 [16]. Characteristics of patients who were censored after 10 yr or had died from PCa within 10 yr were compared with univariate chisquare or Wilcoxon rank sum tests for categorical and continuous variables, respectively. Because the study cohorts incorporated case-control and cohort study designs, survival analysis was not appropriate. Although there are methods for adapting proportional hazards models to case-control data [17], they are not suitable when the data combine both case-control and cohort data. Therefore, we conservatively used a logistic regression approach with PCSM10 as the outcome.
The ability of the GC to improve upon prognostic information in clinical variables was evaluated in two ways. Unconditional logistic regression models were fit to individual clinical variables (PSA, RP Gleason score, RP stage) to generate a base model, then the GC was added to the base model. Alternatively, the GC was added to a model with CAPRA-S as a validated measure of postoperative risk. The latter approach may be a more realistic indication of GC performance because the base clinical model is derived from this dataset, hence subject to overfitting [18], whereas the GC and CAPRA-S were both trained and validated on datasets independent of the current data. Models were fit for: (1) all men with adverse pathologic features and PCSM10 defined, (2) men considered high risk (PSA > 20 ng/ml or RP Gleason score 8–10 or stage pT3b or pN1) [10], and (3) men at very high risk of death due to biochemical recurrence within 2 yr (BCR2) [1] or metastasis (MET). For the overall and subgroup analyses the time frame for PCSM10 began with the date of RP. The univariate and adjusted effect of GC were measured by the odds ratio (OR) and 95% confidence interval for a 0.1 increase in the score (GC ranges from 0 to 1), or for GC high (> 0.6) versus low-intermediate (≤ 0.6). The bootstrap corrected area under the receiver operating characteristic curve (AUC) was used to determine incremental improvement in model performance by adding the GC to CAPRA-S or to the base model. Decision curve analysis was used to compare the net benefit associated with PCSM10 prediction using the base model, GC or CAPRA-S alone, and GC combined with either the base model or CAPRA-S [19]. Although bootstrapping was used for internal validation of the models, performance of the models must be regarded as best case scenario until externally validated [20]. Because the analysis combines data from four institutions, analyses that stratified by institution were also performed. These models gave nearly identical results to the unstratified models so only the latter are reported. Models were fit using R v3.2.0 (R Foundation for Statistical Computing, Vienna, Austria) and SAS v9.4 (SAS Institute, Cary, NC, USA).
3. Results
The analytic cohort consisted of 561 patients either censored alive after 10 yr of follow-up or died of PCa within 10 yr (n = 112). Median follow-up was 13.0 yr in censored patients, 6.0 yr in men with PCSM10, and 12 yr overall. Median GC was 0.39 (interquartile range: 0.23, 0.59), and median CAPRA-S was 4 (interquartile range: 3, 6). Table 1 compares patient characteristics by PCSM10 status while Supplementary Table 1 compares characteristics of the analytic cohort to those of patients excluded with <10 yr of follow-up. As expected, patients with PCSM10 had more adverse characteristics including PSA, RP Gleason score, RP stage, CAPRA-S, and GC score, while patients in the analytic cohort were found to be at slightly higher risk than those excluded with <10 yr of follow-up. Adjuvant therapy (androgen deprivation or radiotherapy), administered to 15% of patients, was associated with a higher risk of PCSM10, reflecting confounding by indication, that is adjuvant treatment given to higher risk patients.
Table 1.
Characteristics of men with adverse pathologic features at prostatectomy who were censored or alive at 10 yr compared with those who died of prostate cancer within 10 yr of surgery
| Variable | No PCSM at ≥10 yr (n = 449) |
PCSM in ≤10 yr (n = 112) |
p valuea |
|---|---|---|---|
|
| |||
| Age, median (IQR) | 62 (57–67) | 61 (57–66) | 0.6 |
|
| |||
| PSA, n (%) | 0.6 | ||
| <10 | 252 (56) | 57 (51) | |
| 10–20 | 124 (28) | 33 (29) | |
| >20 | 73 (16) | 22(20) | |
|
| |||
| RP Gleason, n (%) | <0.001 | ||
| ≤6 | 39 (9) | 0 (0) | |
| 7 | 285 (63) | 32 (29) | |
| 8–10 | 125 (28) | 80 (71) | |
|
| |||
| RP Stage, n (%) | <0.001 | ||
| OC | 168 (37) | 17 (15) | |
| EPE | 147 (33) | 28 (25) | |
| SVI | 87 (19) | 38 (34) | |
| LN | 47 (10) | 29(26) | |
|
| |||
| Surgical margins, n (%) | 0.7 | ||
| Negative | 234 (52) | 61 (54) | |
| Positive | 215 (48) | 51 (46) | |
|
| |||
| CAPRA-S, n (%) | <0.001 | ||
| <3 | 103 (23) | 5 (5) | |
| 3–5 | 200(45) | 36 (32) | |
| >5 | 146 (33) | 71 (63) | |
|
| |||
| GC, n (%) | <0.001 | ||
| <0.45 | 289(64) | 38 (34) | |
| 0.45–0.6 | 84 (19) | 13 (12) | |
| >0.6 | 76 (17) | 61 (54) | |
|
| |||
| Adjuvant ADT or RT, n (%) | <0.001 | ||
| No | 395 (88) | 80 (71) | |
| Yes | 54 (12) | 32 (29) | |
ADT = androgen deprivation therapy; CAPRA-S = Cancer of the Prostate Risk Assessment Postsurgical; EPE = extraprostatic extension; GC = Decipher genomic classifier; IQR = interquartile range; LNI = lymph node involvement; OC = organ confined; PCSM = prostate cancer-specific mortality; PSA = prostate-specific antigen; RP = radical prostatectomy; RT = radiation therapy; SVI = seminal vesicle involvement.
Wilcoxon rank sum, Pearson chi-square, or Mantel-Haenszel chi-square test.
To evaluate how addition of the dichotomous GC score could further stratify risk of PCSM at 10 yr within CAPRA-S categories we excluded the case-control data from analysis to allow calculation of cumulative incidence, adjusted for competing risk due to other cause deaths (further details in Supplementary data). Jointly stratifying risk by dichotomous GC score ≤0.6 (low-intermediate) versus >0.6 (high) and CAPRA-S <6 (low-intermediate) versus ≥6 (high) revealed a wide range of risk within CAPRA-S categories. Among patients with low-intermediate risk CAPRA-S, the GC further stratifies PCSM10 risk from 2.8% for GC ≤0.6 to 18% for GC >0.6. Among high risk CAPRA-S patients GC stratifies risk from 5.5% for GC ≤0.6 to 30% for GC >0.6. Addition of the GC provides a nearly 6-fold stratification of risk within CAPRA-S categories. Tables 2–6 demonstrate the association of GC with risk of PCSM10 from univariate logistic regression models, and models adjusted for base clinical variables, or for CAPRA-S (continuous). Models were fit to all patients in the analytic cohort (n=561, 112 PCSM10), only those with high risk (PSA > 20 ng/ml or RPRP Gleason score 8–10 or RP stage pT3b or N1, n = 323, 98 PCSM10), and very high risk patients with either BCR2 (n = 212, 86 PCSM10), or with metastasis (MET, n = 230, 112 PCSM10), and patients without postoperative treatment (n = 475, 80 PCSM10).
Table 2.
PCSM10 risk associated with GC (per 0.1 unit or dichotomized at ≥ 0.6 vs ≤ 0.6) in logistic regression models: univariate GC and GC adjusted for base clinical model or CAPRA-S (all patients [n = 561, 112 with PCSM10])
| Model | OR (95% CI) | p value | AUC of model (95% CI) | Increase in AUC from adding GC |
|---|---|---|---|---|
| GC (per 0.1 unit) | 1.48 (1.33, 1.64) | <0.001 | 0.73 (0.67, 0.78) | (NA) |
| CAPRA-S (per 1 unit) | 1.38 (1.27, 1.51) | <0.001 | 0.73 (0.68, 0.78) | (NA) |
| Base model | (NA)* | <0.001 | 0.77 (0.73, 0.83) | (NA) |
| GC (per 0.1 unit), adjusted for base model | 1.32 (1.19, 1.48) | <0.001 | 0.79 (0.75, 0.84) | (0.03) |
| GC (per 0.1 unit), adjusted for CAPRA-S | 1.34 (1.20, 1.50) | <0.001 | 0.76 (0.71, 0.82) | (0.03) |
| GC (> 0.6 vs ≤ 0.6) | 5.87 (3.76, 9.17) | <0.001 | 0.69 (0.64, 0.74) | (NA) |
| GC (> 0.6 vs ≤ 0.6), adjusted forbase model | 3.87 (2.36, 6.35) | <0.001 | 0.80 (0.76, 0.85) | (0.04) |
| GC (> 0.6 vs ≤ 0.6), adjusted for CAPRA-S | 3.91 (2.43, 6.29) | <0.001 | 0.77 (0.77, 0.81) | (0.04) |
AUC = area under the curve; CAPRA-S = Cancer of the Prostate Risk Assessment Postsurgical; CI = confidence interval; GC = Decipher genomic classifier; NA = not applicable; OR = odds ratio; PCSM = prostate cancer-specific mortality.
The base model includes preoperative prostate-specific antigen (continuous), prostatectomy Gleason score (7, 8, 9–10), prostatectomy stage (organ confined, extraprostatic extension, seminal vesicle involvement, lymph node metastasis). Because these are well established prognostic factors but are not the focus of this study, the individual odds ratios and 95% confidence intervals for these variables are not shown.
Table 6.
Patients without post-operative treatment (n = 475, 80 with PCSM10)
| Model | OR (95% CI) | p value | AUC of model (95% CI) | Increase in AUC from adding GC |
|---|---|---|---|---|
| GC (per 0.1 unit) | 1.36 (1.22, 1.53) | <0.001 | 0.68 (0.61, 0.75) | (NA) |
| CAPRA-S (per 1 unit) | 1.42 (1.26, 1.59) | <0.001 | 0.71 (0.66, 0.77) | (NA) |
| Base model | (NA)a | <0.001 | 0.76 (0.70, 0.82) | (NA) |
| GC (per 0.1 unit), adjusted for base model | 1.20 (1.06, 1.37) | 0.005 | 0.78 (0.72, 0.83) | (0.02) |
| GC (per 0.1 unit), adjusted for CAPRA-S | 1.26 (1.12, 1.43) | <0.001 | 0.74 (0.68, 0.80) | (0.03) |
| GC (> 0.6 vs ≤ 0.6) | 4.48 (2.67, 7.52) | <0.001 | 0.58 (0.59, 0.71) | (NA) |
| GC (> 0.6 vs ≤ 0.6), adjusted for base model | 2.87 (1.62, 5.10) | <0.001 | 0.78 (0.73, 0.84) | (0.02) |
| GC (> 0.6 vs ≤ 0.6), adjusted for CAPRA-S | 3.29 (1.90, 5.67) | <0.001 | 0.74 (0.69, 0.80) | (0.03) |
AUC = area under the curve; CAPRA-S = Cancer of the Prostate Risk Assessment Postsurgical; CI = confidence interval; GC = Decipher genomic classifier; NA = not applicable; OR = odds ratio; PCSM = prostate cancer-specific mortality.
The base model includes preoperative prostate-specific antigen (continuous), prostatectomy Gleason score (7, 8, 9–10), prostatectomy stage (organ confined, extraprostatic extension, seminal vesicle involvement, lymph node metastasis). Because these are well established prognostic factors but are not the focus of this study, the individual odds ratios and 95% confidence intervals for these variables are not shown.
Among all patients, high GC score (> 0.6) was associated with a statistically significant increase in risk of PCSM10 after adjusting for CAPRA-S, OR = 3.91, and AUC = 0.77 (increase in AUC of 0.04 over model with CAPRA-S alone; Table 2). Figure 1 compares the AUC for each fit of the models to all patients. Figure 2 compares decision curves among the models for all patients. For thresholds above 20% the GC yields higher net benefit than CAPRA-S, and the combination of GC+CAPRA-S yields higher net benefit than either classifier alone. As mentioned in Section 2, performance of the base clinical variable model (PSA, RP Gleason score, RP stage) is subject to bias due to over-fitting to this dataset.
Fig. 1. Area under the receiver operating characteristic curve for the models of prostate cancer-specific mortality within 10 yr in all patients.
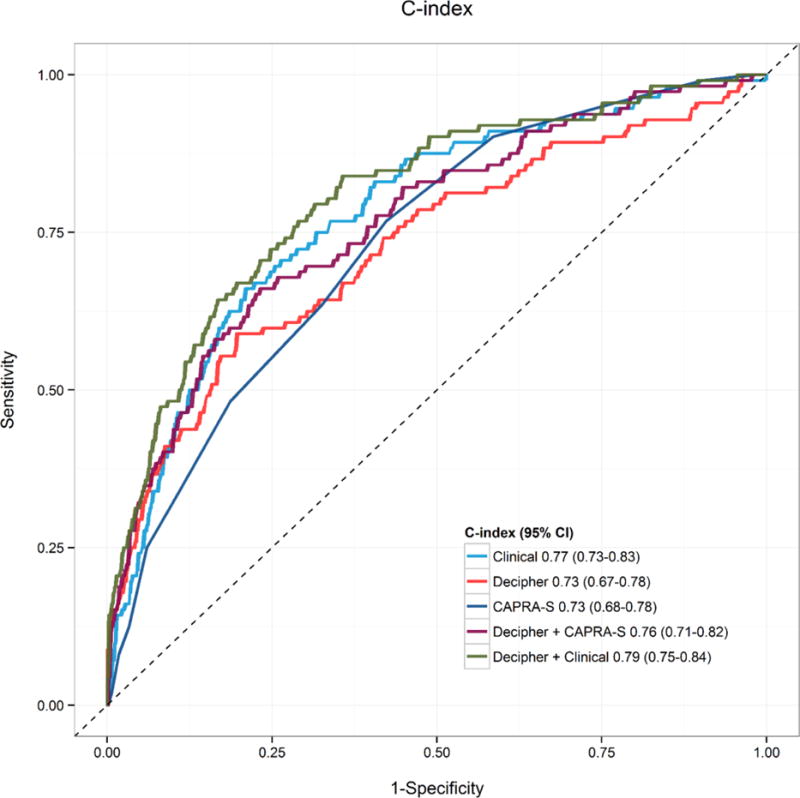
CAPRA-S = Cancer of the Prostate Risk Assessment Postsurgical Score; CI = confidence interval.
Fig. 2. Decision curve analysis for the models of prostate cancer-specific mortality within 10 yr in all patients.
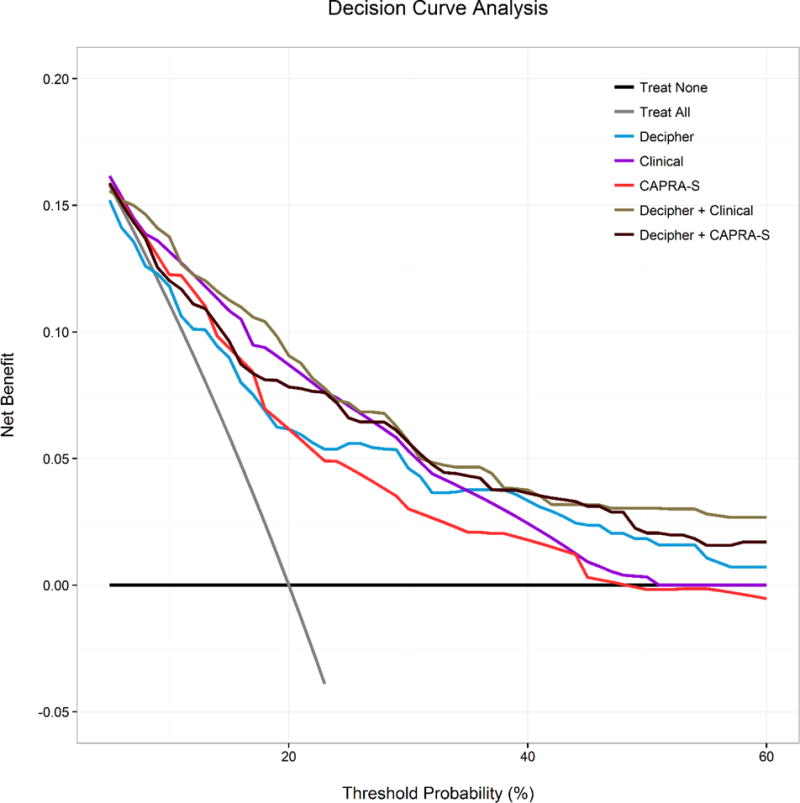
CAPRA-S = Cancer of the Prostate Risk Assessment Postsurgical Score.
High GC score also performed well in the high and very high risk subsets: high risk patients, OR = 3.96, AUC = 0.69 (increase 0.08 over CAPRA-S); BCR2, OR = 3.06, AUC = 0.72 (increase 0.03 over CAPRA-S); MET, OR = 1.95, AUC = 0.64 (increase 0.03 over CAPRA-S; Tables 3–6). Univariate results for the base model and CAPRA-S are in Supplementary Table 3. Adjustment for clinical variables or CAPRA-S causes only small decreases to the OR for GC, suggesting that the GC captures important prognostic information that is independent of clinical variables. In contrast, CAPRA-S as a continuous variable was not statistically significant after adjustment for GC among high risk patients, and when dichotomized as > 6 versus <6 [10], it was not statistically significant when adjusting for GC among high risk, BCR2, and MET patients (Supplementary Table 3).
Table 3.
Patients with high risk (PSA > 20 or prostatectomy Gleason score > 8 or prostatectomy stage pT3b/N1 [n = 323, 98 PCSM10]
| Model | OR (95% CI) | p value | AUC of model (95% CI) | Increase in AUC from adding GC |
|---|---|---|---|---|
| GC (per 0.1 unit) | 1.37 (1.21, 1.54) | <0.001 | 0.69 (0.62, 0.76) | (NA) |
| GC (per 0.1 unit), adjusted for base model | 1.31 (1.15, 1.48) | <0.001 | 0.73 (0.67, 0.78) | (0.04) |
| GC (per 0.1 unit), adjusted for CAPRA-S | 1.33 (1.17, 1.50) | <0.001 | 0.69 (0.62, 0.75) | (0.08) |
| GC (>0.6 vs. ≤0.6) | 4.51 (2.72, 7.48) | <0.001 | 0.67 (0.62, 0.73) | (NA) |
| GC (>0.6 vs. ≤0.6) + base model | 3.90 (2.27,6.70) | <0.001 | 0.74 (0.69, 0.80) | (0.06) |
| GC (>0.6 vs. ≤0.6) + CAPRA-S | 3.96 (2.35, 6.69) | <0.001 | 0.69 (0.63, 0.76) | (0.08) |
AUC = area under the curve; CAPRA-S = Cancer of the Prostate Risk Assessment Postsurgical; CI = confidence interval; GC = Decipher genomic classifier; NA = not applicable; OR = odds ratio; PCSM = prostate cancer-specific mortality; PSA = prostate-specific antigen.
When patients with adjuvant therapy were excluded from analyses the results were comparable to those of the overall analysis (Table 6). Results were also similar when adjusting for adjuvant therapy, and interactions between GC and adjuvant therapy were not significant in any of the risk groups (data not shown). Exclusion of case-control patients to allow proportional hazards survival analysis also yielded similar results (Supplementary data, Supplementary Table 4).
4. Discussion
Despite the strong prognostic value of Gleason score, stage, and PSA in men who have undergone RP, there remains considerable variability in risk of dying from, rather than with PCa. Even among men with adverse pathologic features or who have experienced BCR, the majority will not die of their disease [1,4,21,22]. This has contributed to difficulty in developing clear-cut guidelines and algorithms to select patients most likely to benefit from adjuvant or salvage therapy [23,24]. The problem is exacerbated now that new treatments have been shown to extend life in the metastatic setting, creating interest in earlier sequencing of novel therapeutic combinations.
The current study is the first large multi-institutional study of men with adverse pathologic features to demonstrate that Decipher GC can provide additional stratification of PCSM risk beyond clinical variables or CAPRA-S alone, and provides independent prognostic information even among subgroups of men at high risk according to clinical and pathology features, or very high risk due to early BCR or development of metastases. Previous multi-institutional validations of the GC have shown it to independently predict metastasis risk among men with intermediate to high risk after RP [9,11,25]. A genomic-clinical classifier that integrated the GC with CAPRA-S improved PCSM prediction compared to either classifier alone in a small single institution cohort of 185 high risk men with only 28 PCa deaths [10].
In the current study stratifying by both CAPRA-S and GC revealed a range of PCSM10 risk from 2.8% to 30%. In fact, among CAPRA-S low patients 11% had a high-risk GC score, while among CAPRA-S high patients, 41% had a low risk GC score. These two extreme shifts in risk comprised 18% of the total study sample, and give an estimate of the potential fraction of intermediate-high risk patients whose clinical management may be changed by GC combined with CAPRA-S. It is encouraging that the subgroup classified as low-intermediate by both GC and CAPRA-S comprises 53% of the sample, indicating that a substantial number of men with adverse pathologic features have a low risk of dying from PCa. Additionally, men classified high risk by CAPRA-S but low risk by GC may represent a subgroup who do not require postsurgical therapy until evidence of systemic disease progression. These findings validate results observed in the small single institution study, from which 52 patients overlap the current analysis cohort [10]. To our knowledge only two other genomic signatures have been evaluated as predictors of PCSM in multivariable models adjusted for clinical and pathologic prognostic factors. However, these signatures either were evaluated among conservatively managed patients most of whom were at high enough risk that they would undergo primary therapy by contemporary practice [26], or the signatures were trained and tested in the same patients without undergoing independent validation [27]. Cuzick et al [26] validated a previously developed cell cycle proliferation signature, in a cohort with 40% of men having biopsy Gleason score ≥4+3 and 68% PSA >10 ng/ml. Some men eventually received treatment but this information was known for only a minority of patients and could not be used in analysis. The only other signature evaluated for PCSM in a RP cohort included men at intermediate-high risk. This study developed a proteomic signature that was an independent predictor of PCSM; however, this signature has not been validated. Furthermore, although the classifier was associated with a very large hazard ratio for PCSM per 1 unit change in the risk score, the distribution of risk scores was not provided so it is unclear whether a 1 unit change represents a clinically realistic metric [27]. The current validation of the Decipher GC as a predictor of PCSM has a number of strengths. The GC has been validated as a predictor of metastasis in independent cohorts [6–9,11]. The current study was larger than any of the other studies of classifiers and PCSM except for the study by Cuzick et al [26], and had a larger number of deaths than any other study. The GC was also shown to be independently predictive of PCSM even in subgroups at high to very high risk, and when combined with CAPRA-S, provided a wide stratification of risk. It is also noteworthy that, despite clonal evoluation separating primary from lethal metastatic tumors, the primary tumor contains important biological signal that persists in the lethal phenotype and is captured by Decipher.
There are also some limitations. We used PCSM within 10 yr as an endpoint rather than survival time because the data set combined case-control and case-cohort study designs. Twenty percent of PCa deaths occurred later than 10 yr; including them among the controls may have attenuated the risk associated with high GC scores. However, we performed a survival analysis after excluding case-control patients, and the results were similar to the overall analysis of PCSM10. Furthermore, the hazard ratio for a full cohort study is appropriately estimated by the OR from logistic regression analysis of a case-control study, albeit with a larger standard error, so our approach is likely to be conservative[17,28,29]. CAPRA-S was not originally developed in a population of largely high-risk patients, so there is the possibility that it underperforms in the current study, although the OR = 1.38, similar to the hazard ratio = 1.42 for PCSM in Cooperberg et al’s original paper [16]. Adjuvant therapy practice differed among the four institutions, and was administered to only 15% of patients, which is comparable to national trends [30]. Although this number was too small for a subgroup analysis, the effect of GC did not change when adjusted for adjuvant therapy, and results also did not change if adjuvant therapy patients were excluded from analysis. Lastly, our patient population was largely from tertiary academic centers, which may affect the generalizability of our results. However, the proportional shifts from CAPRA-S low risk to GC high risk, and vice versa, were comparable to those seen in a study of the GC impact on clinical decision-making among urologists, 55% of whom were in community practice [31].
5. Conclusions
In a large multicenter sample of men with adverse pathologic features with the longest follow-up to date, the Decipher GC demonstrated a potentially clinically important improvement in prediction of PCSM at 10 yr in all men, as well as in subgroups considered high risk based on PSA, stage. or Gleason score, and very high risk men who experienced early BCR, or who developed metastasis after RP. Adding the GC to a model with either CAPRA-S or individual clinical variables improved prediction of PCSM10, and identified subgroups of men with very low risk of PCSM at 10 yr (10%) who may be suitable for conservative postsurgical management, and men with very high risk (55%) who may require more aggressive therapy. Clinical trials are needed to determine whether the combination of the Decipher GC and CAPRA-S can improve treatment assignment and outcomes compared to clinical variables alone.
In men with adverse pathology or early disease progression after prostatectomy, the Decipher 22 gene genomic classifier predicts risk of prostate cancer death. When combined with the Cancer of the Prostate Risk Assessment Postsurgical Score it further stratifies risk, which may be useful for decisions about postprostatectomy treatment.
Supplementary Material
Table 4.
Patients with BCR within 2 yr (n = 212, 86 with PCSM10)
| Model | OR (95% CI) | p value | AUC of model (95% CI) | Increase in AUC from adding GC |
|---|---|---|---|---|
| GC (per 0.1 unit) | 1.35 (1.18, 1.53) | <0.001 | 0.69 (0.62, 0.77) | (NA) |
| GC (per 0.1 unit), adjusted for base model | 1.26 (1.09, 1.45) | 0.002 | 0.73 (0.67, 0.80) | (0.03) |
| GC (per 0.1 unit), adjusted for CAPRA-S | 1.25 (1.09, 1.43) | 0.002 | 0.71 (0.63, 0.78) | (0.02) |
| GC (> 0.6 vs ≤ 0.6) | 4.22 (2.32, 7.66) | <0.001 | 0.66 (0.60, 0.73) | (NA) |
| GC (> 0.6 vs ≤ 0.6), adjusted for base model | 3.49 (1.80, 6.77) | <0.001 | 0.75 (0.68, 0.81) | (0.04) |
| GC (> 0.6 vs ≤ 0.6), adjusted for CAPRA-S | 3.06 (1.62, 5.76) | <0.001 | 0.72 (0.65, 0.79) | (0.03) |
AUC = area under the curve; BCR = biochemical recurrence; CAPRA-S = Cancer of the Prostate Risk Assessment Postsurgical; CI = confidence interval; GC = Decipher genomic classifier; NA = not applicable; OR = odds ratio; PCSM = prostate cancer-specific mortality.
Table 5.
Patients with metastasis (n = 230, 112 with PCSM10)
| Model | OR (95% CI) | p value | AUC of model (95% CI) | Increase in AUC from adding GC |
|---|---|---|---|---|
| GC (per 0.1 unit) | 1.19 (1.06, 1.34) | 0.003 | 0.63 (0.55, 0.70) | (NA) |
| GC (per 0.1 unit), adjusted for base model | 1.19 (1.05, 1.34) | 0.006 | 0.63 (0.56, 0.70) | (0.04) |
| GC (per 0.1 unit), adjusted for CAPRA-S | 1.14 (1.01, 1.29) | 0.03 | 0.64 (0.56, 0.70) | (0.03) |
| GC (> 0.6 vs ≤ 0.6) | 2.33 (1.37, 3.97) | 0.002 | 0.60 (0.54, 0.67) | (NA) |
| GC (> 0.6 vs ≤ 0.6) + base model | 2.31 (1.31, 4.06) | 0.004 | 0.63 (0.56, 0.70) | (0.04) |
| GC (> 0.6 vs ≤ 0.6) + CAPRA-S | 1.95 (1.12, 3.39) | 0.02 | 0.64 (0.56, 0.71) | (0.03) |
AUC = area under the curve; CAPRA-S = Cancer of the Prostate Risk Assessment Postsurgical; CI = confidence interval; GC = Decipher genomic classifier; NA = not applicable; OR = odds ratio; PCSM = prostate cancer-specific mortality.
Acknowledgments
Funding/Support and role of the sponsor: DOD/PCRP W81XWH-14-2-0182 Prostate Cancer Biorepository Network; NIH P50 CA058236 Johns Hopkins Prostate Cancer SPORE grant; GenomeDx Biosciences, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: Bruce J. Trock had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Trock.
Acquisition of data: Karnes, Ross, Schaeffer, Klein, Freedland, Takhar.
Analysis and interpretation of data: Trock, Choeurng, Davicioni.
Drafting of the manuscript: Trock.
Critical revision of the manuscript for important intellectual content: Trock, Karnes, Ross, Schaeffer, Klein, Freedland, Cooperberg.
Statistical analysis: Trock, Choeurng, Yousefi.
Obtaining funding: Trock, Karnes, Schaeffer, Davicioni.
Administrative, technical, or material support: Erho, Takhar.
Supervision: Trock, Davicioni.
Other: None.
Financial disclosures: Bruce J. Trock certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Trock has been a consultant to GenomeDx Biosciences, and has received research grant support from Myriad Genetics, Inc.; Ross, Schaeffer, and Cooperberg have been consultants to GenomeDx Biosciences; Choeurng, Erho, Yousefi, Takhar, and Davicioni are employees of GenomeDx Biosciences.
References
- 1.Freedland SJ, Humphreys EB, Mangold LA, et al. Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. JAMA. 2005;294:433–9. doi: 10.1001/jama.294.4.433. [DOI] [PubMed] [Google Scholar]
- 2.Miyahira AK, Lang JM, Den RB, et al. Multidisciplinary intervention of early, lethal metastatic prostate cancer: Report from the 2015 Coffey-Holden Prostate Cancer Academy Meeting. Prostate. 2016;76:125–39. doi: 10.1002/pros.23107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cha EK, Eastham JA. Chemotherapy and novel therapeutics before radical prostatectomy for high-risk clinically localized prostate cancer. Urol Oncol. 2015;33:217–25. doi: 10.1016/j.urolonc.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 4.Eggener SE, Scardino PT, Walsh PC, et al. Predicting 15-year prostate cancer specific mortality after radical prostatectomy. J Urol. 2011;185:869–75. doi: 10.1016/j.juro.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Punnen S, Freedland SJ, Presti JC, et al. Multi-institutional validation of the CAPRA-S score to predict disease recurrence and mortality after radical prostatectomy. Eur Urol. 2014;65:1171–7. doi: 10.1016/j.eururo.2013.03.058. [DOI] [PubMed] [Google Scholar]
- 6.Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeffrey Karnes R, Bergstralh EJ, Davicioni E, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk Patient population. J Urol. 2013;190:2047–53. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein EA, Haddad Z, Yousefi K, et al. Decipher genomic classifier measured on prostate biopsy predicts metastasis risk. Urology. 2016;90:148–52. doi: 10.1016/j.urology.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Ross AE, Feng FY, Ghadessi M, et al. A genomic classifier predicting metastatic disease progression in men with biochemical recurrence after prostatectomy. Prostate Cancer Prostatic Dis. 2014;17:64–9. doi: 10.1038/pcan.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooperberg MR, Davicioni E, Crisan A, Jenkins RB, Ghadessi M, Karnes RJ. Combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high-risk prostatectomy cohort. Eur Urol. 2015;67:326–33. doi: 10.1016/j.eururo.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein EA, Yousefi K, Haddad Z, et al. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur Urol. 2015;67:778–86. doi: 10.1016/j.eururo.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 12.Ross AE, Johnson MH, Yousefi K, et al. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate-and high-risk men. Eur Urol. 2016;69:157–65. doi: 10.1016/j.eururo.2015.05.042. [DOI] [PubMed] [Google Scholar]
- 13.Freedland SJ, Choeurng V, Howard L, et al. Utilization of a genomic classifier for prediction of metastasis following salvage radiation therapy after radical prostatectomy. Eur Urol. 2016;70:588–96. doi: 10.1016/j.eururo.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Lockstone HE. Exon array data analysis using Affymetrix power tools and R statistical software. Brief Bioinform. 2011;12:634–44. doi: 10.1093/bib/bbq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piccolo SR, Sun Y, Campbell JD, Lenburg ME, Bild AH, Johnson WE. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics. 2012;100:337–44. doi: 10.1016/j.ygeno.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooperberg MR, Hilton JF, Carroll PR. The CAPRA-S score: A straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer. 2011;117:5039–46. doi: 10.1002/cncr.26169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prentice Rl, Breslow NE. Retrospective studies and failure time models. Biometrika. 1978;65:153–8. [Google Scholar]
- 18.Harrell FE, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–87. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Mak. 2006;26:565–74. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steyerberg EW, Harrell FE, Jr, Borsboom GJJM, Eijkemans MJC, Vergouwe Y, Habbema JDF. Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. J Clin Epidemiol. 2001;54:774–81. doi: 10.1016/s0895-4356(01)00341-9. [DOI] [PubMed] [Google Scholar]
- 21.Xia J, Trock BJ, Gulati R, et al. Overdetection of recurrence after radical prostatectomy: estimates based on patient and tumor characteristics. Clin Cancer Res. 2014;20:5302–10. doi: 10.1158/1078-0432.CCR-13-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boorjian SA, Thompson RH, Tollefson MK, et al. Long-term risk of clinical progression after biochemical recurrence following radical prostatectomy: the impact of time from surgery to recurrence. Eur Urol. 2011;59:893–9. doi: 10.1016/j.eururo.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 23.Falchook AD, Chen RC. Adjuvant vs. salvage radiotherapy for patients at high risk for recurrence after radical prostatectomy. Urol Oncol. 2015;33:451–5. doi: 10.1016/j.urolonc.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 24.Fossati N, Karnes RJ, Boorjian SA, et al. Long-term impact of adjuvant versus early salvage radiation therapy in pT3N0 prostate cancer patients treated with radical prostatectomy: results from a multi-institutional series. Eur Urol. doi: 10.1016/j.eururo.2016.07.028. http://dx.doi.org/10.1016/j.eururo.2016.07.028. [DOI] [PubMed]
- 25.Freedland SJ, Choeurng V, Howard L, De Hoedt A. Utilization of a genomic classifier for prediction of metastasis following salvage radiation therapy after radical prostatectomy. Eur Urol. 2016;70:588–96. doi: 10.1016/j.eururo.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Cuzick J, Stone S, Fisher G, et al. Validation of an RNA cell cycle progression score for predicting death from prostate cancer in a conservatively managed needle biopsy cohort. Br J Cancer. 2015;113:382–9. doi: 10.1038/bjc.2015.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shipitsin M, Small C, Choudhury S, et al. Identification of proteomic biomarkers predicting prostate cancer aggressiveness and lethality despite biopsy-sampling error. Br J Cancer. 2014;111:1201–12. doi: 10.1038/bjc.2014.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalbfleisch JD, Prentice RL. The statistical analysis of failure-time data. New York, NY: John Wiley and Sons; 1980. [Google Scholar]
- 29.Whittemore AS. Efficiency of synthetic retrospective studies. Biom J. 1981;23:73–8. [Google Scholar]
- 30.Sineshaw HM, Gray PJ, Efstathiou JA, Jemal A. Declining use of radiotherapy for adverse features after radical prostatectomy: results from the National Cancer Data Base. Eur Urol. 2015;68:768–74. doi: 10.1016/j.eururo.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Badani KK, Thompson DJ, Brown G, et al. Effect of a genomic classifier test on clinical practice decisions for patients with high risk prostate cancer after surgery. BJU Int. 2015;115:419–29. doi: 10.1111/bju.12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


