Table 1.
Identification of conserved miRNA isoforms identified in both Arabidopsis and P. glauca during seed set.
| Namea | Predicted targetb | Alignment | Target gene functionc | Ed | UPEe | |
|---|---|---|---|---|---|---|
| miR159b-3p | AT1G18080.1 | miRNA |  |
GA and flowering pathways | 0 | 22 |
| Target | ||||||
| BT123375 | miRNA | 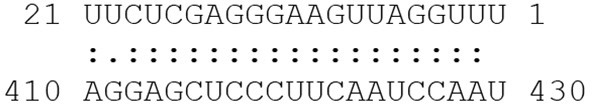 |
– | 1.5 | 12 | |
| Target | ||||||
| miR160c-5p | AT1G77850 AT2G28350 AT4G30080 | miRNA | 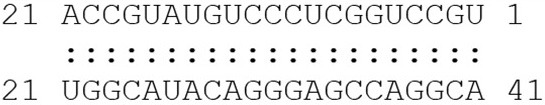 |
AUXIN RESPONSE FACTOR (ARF) 10, 16 and 17 | 0 | 17 |
| Target | ||||||
| BT119832 | miRNA | 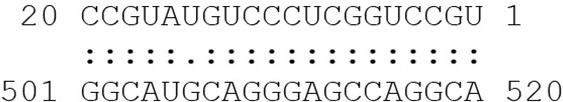 |
Putative ARF 10/16/17 (NCBI No. FN433183) in Cycas rumphii | 0.8 | 19 | |
| Target | ||||||
| miR163 | AT1G66720.1 | miRNA | 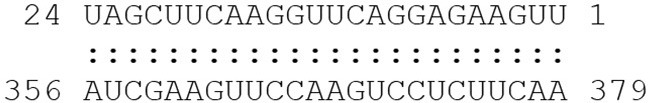 |
Methylation | 0 | 13 |
| Target | ||||||
| BT112171 | miRNA |  |
– | 2.5 | 15 | |
| Target | ||||||
| miR166b-5p | AT4G14713.1 | miRNA |  |
Cell proliferation | 2 | 13 |
| Target | ||||||
| EF677221 | miRNA | 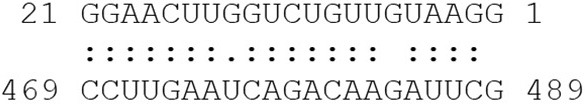 |
– | 3 | 15 | |
| Target | ||||||
| miR166g | AT2G34710 | miRNA | 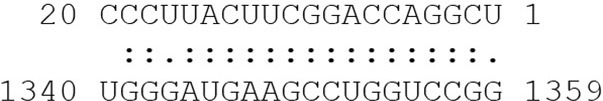 |
HOMEOBOX PROTEIN 14, associated with development | 2 | 21 |
| Target | ||||||
| HQ391915 | miRNA | 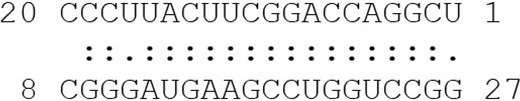 |
Homeodomain leucine zipper protein | 2 | 16 | |
| Target | ||||||
| miR167a-5p | AT1G30330 AT5G37020 | miRNA | 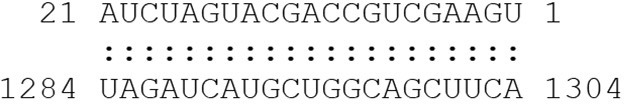 |
AUXIN RESPONSE FACTOR 6 and 8 | 0 | 24 |
| Target | ||||||
| FJ469921 | miRNA |  |
R2R3-MYB transcription factor | 3 | 18 | |
| Target | ||||||
| miR171a-3p | AT3G60630.1 | miRNA | 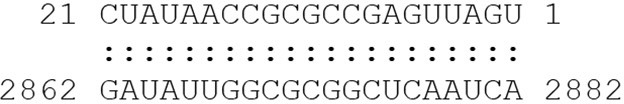 |
Cell differentiation and division | 0 | 14 |
| Target | ||||||
| BT102743 | miRNA |  |
– | 0 | 17 | |
| Target | ||||||
| miR319b | AT4G23710.1 | miRNA |  |
Proton transport | 0 | 15 |
| Target | ||||||
| BT110042.1 | miRNA | 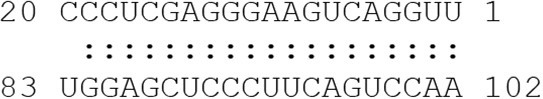 |
– | 1 | 22 | |
| Target | ||||||
| miR390b-5p | AT5G03650.1 | miRNA | 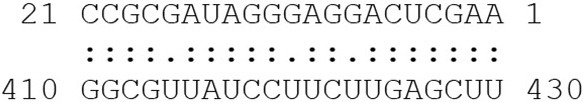 |
Starch branching enzyme | 1.5 | 21 |
| Target | ||||||
| EX354481 | miRNA | 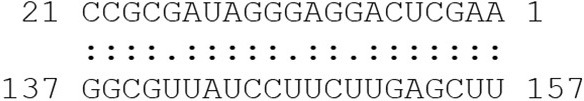 |
– | 1.5 | 17 | |
| Target | ||||||
| miR394b-5p | AT1G27350 | miRNA | 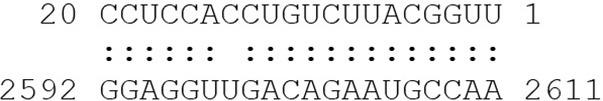 |
Ribosome associated membrane protein | 1 | 15 |
| Target | ||||||
| BT112917 | miRNA | 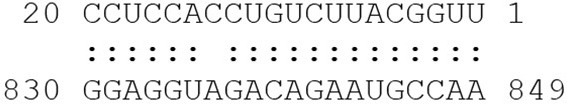 |
– | 1 | 20 | |
| Target | ||||||
| miR396-5p | AT1G53910 | miRNA | 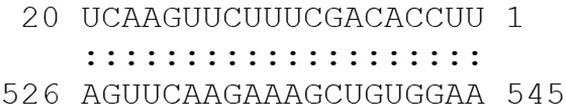 |
Ethylene response factor | 0 | 14 |
| Target | ||||||
| BT102125 | miRNA | 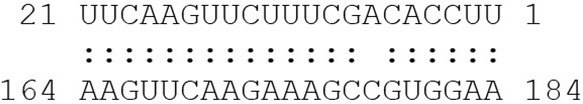 |
– | 1.5 | 34 | |
| Target | ||||||
| miR408-3p | AT2G02860 | miRNA |  |
SUCROSE TRANSPORTER 3 | 1 | 23 |
| Target | ||||||
| BT103532 | miRNA | 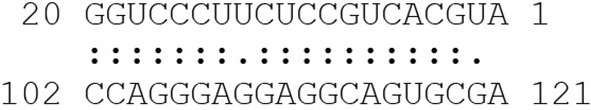 |
– | 2 | 25 | |
| Target | ||||||
| miR824-5p | AT3G57230 | miRNA |  |
MADS-box transcription factor | 0.5 | 15 |
| Target | ||||||
| BT112142 | miRNA |  |
– | 3 | 13 | |
| Target | ||||||
Nomenclature of miR: (organism)miRnx - precursor arm and/ or.y, where n, a sequential number representing family of miR; x, lettered suffixes representing family member (i.e., closely related mature sequences); −5p or −3p denote 5′ or 3′ arm of the precursor;.y, integer denoting occurrence of more than one mature sequence from the same precursor.
For each miRNA, shown is the most confidently predicted in Arabidopsis (above) and P. glauca (below).
Refer to GO enrichment analysis, TAIR10 and NCBI.
Expectation (E), stringent threshold [0–0.2] gives lower false positive prediction.
Maximum energy to unpair the target site (UPE), small value (range ϵ [0, 100]) is better.
