Abstract
Retinoic acid (RA), a metabolite of vitamin A, has been found to influence regeneration in the adult central nervous system (CNS). There may be an effect of RA in the recovery/repair in multiple sclerosis (MS), an autoimmune and neurodegenerative disease of the CNS. We hypothesized that RA is a regulator of the further differentiation of oligodendrocyte precursor cells (OPCs) – cells key to the remyelination process in MS. We conducted studies utilizing RNA-sequencing in human embryonic stem cell (hESC)-derived neural stem cells (NSCs) and OPCs so as to understand the role of transcriptional regulators during transition from both ESCs to NSCs and NSCs to OPCs. We identified that expression of retinoic acid receptors β and γ (RARβ and RARγ ) was significantly increased following the transition from NSCs to OPCs. We also demonstrated that long term in vitro culture of hESC-derived OPC with different isoforms of RA led to the significant up-regulation of two known transcriptional inhibitors of oligodendrocyte differentiation: Hes5 following prolonged treatment with all-trans-RA, 9-cis RA and 13-cis RA; and Id4 following treatment with 13cisRA. These results suggest that long term exposure to certain RA isoforms may impact the continued differentiation of this population.
Keywords: Retinoic acid, Retinoic acid receptor beta, Retinoic acid receptor gamma, Neural stem cell, Oligodendrocyte precursor cell
Highlights
-
•
Retinoic acid (ATRA) might have an effect on generation of oligodendrocyte precursor cells (OPC) in the CNS.
-
•
RNA-sequencing used for identification of up-regulation of RARβ and RARɣ expression during OPC differentiation.
-
•
Activation of RARβ and RARɣ by using three different agonists led increase expression of Hes5 and Id4.
1. Introduction
Human neurodevelopment is a highly complex process with many unanswered questions. The study of human embryonic stem cells (hESCs) has contributed greatly to our knowledge of human neural differentiation and neurodevelopment. There have been many studies focusing on mRNA expression during NSC differentiation into cells of the glial cell lineage, and using DNA microarray researchers have been able to assess gene expression differences between NSC and oligodendrocyte precursor cells (OPCs) [1]. However, the transcriptional regulators that drive the lineage fate of human NSCs into human OPCs still requires further investigation.
OPCs represent a large population of adult central nervous system (CNS) resident cells that are capable of proliferation, migration and differentiation [2]. It is postulated that in the adult CNS their primary function is to replace dysfunctional or damaged myelinating oligodendrocytes either after CNS injury or possibly as part of routine CNS homeostasis [3]. Multiple sclerosis (MS) is a chronic autoimmune and neurodegenerative disease of the CNS that is defined pathologically by leukocyte infiltration into the CNS followed by targeted myelin destruction leading to demyelination (lesion) and eventual axonal loss [4]. Key to the prevention of neurological decline is the repair of the CNS that comes in the form of remyelination, for which OPCs are the definitive component [5]. In order for this process to be successful, OPCs need to be recruited to lesion areas where they differentiate into oligodendrocytes and generate new myelin [6]. However, in MS this process appears to fail over time and though a plethora of reasons for failure of remyelination have been suggested, there is a finality to this, with the eventual depletion of OPCs in the lesion [7], [8]. As a potential solution to this problem, approaches have been proposed including the use of stem cell replacement therapy by transplantation of NSCs or OPCs into the CNS or by targeting the production of endogenous new born OPC using RA [9], [10].
Retinoic acid (RA) signaling has many functions within biological systems including a role in the repair of the immune system and the CNS [11], [12]. Receptor engagement by RA initiates a transcriptional cascade that induces further gene expression [13]. Since 9-cis retinoic acid (9cRA) appears to be involved in accelerating CNS myelination via RXRγ signaling in rodents [10], we hypothesized that a function of RA might drive further differentiation of OPCs into mature oligodendrocytes. In this study, we examined differential gene expression by ESCs, NSCs and OPCs using RNA-sequencing (RNA-seq) and describe the expression of RAR (a, β and γ) and RXR (a, β and γ) in these cell populations. We also assessed the effect of RA exposure on these cells and demonstrate that long-term exposure in vitro is associated with an increase in the expression of inhibitory regulators in human OPCs.
2. Material and methods
2.1. Cell culture
Human embryonic stem cells (WA-09) were obtained from University of Southern California Human Stem Cell Core and commercial human NSCs, derived from ESC line H9, were purchased from Invitrogen, respectively. Cellular characteristics and culture conditions have been described previously [14]. Human OPCs were differentiated from ESCs or NSCs as previously described [15], [16].
2.2. OPC sorting and RA treatment
In order the achieve population purity cells were sorted for PDGFRα expression using Flow cytometry (BD FACSAria I cell sorter, BD Bioscience) with anti-PDGFRα (CD140a) PE antibody (BD Pharmingen) [17]. Prior to sorting expression of lineage specific OPC markers were determined by immunohistochemistry (Fig. 2) and confirmed by FACS (data not shown). Sorted cells were analyzed only by FACS and were always >98% pure (data not shown). Following sorting, PDGFRα-positive OPCs were incubated for 24 h in defined culture medium, harvested, and RNA extracted for use in RNA-sequencing. In order to identify the influence of increased RARβ and RARγ expression, 1 μM of all trans-retinoic acid (ATRA), 9-cis retinoic acid (9cRA) or 13-cis retinoic acid (13cRA) (all Sigma) was added every day to human ESC-derived OPCs. After harvesting cells at day 8, 27 and 47, RNA was extracted and cDNA synthesized for RT-PCR.
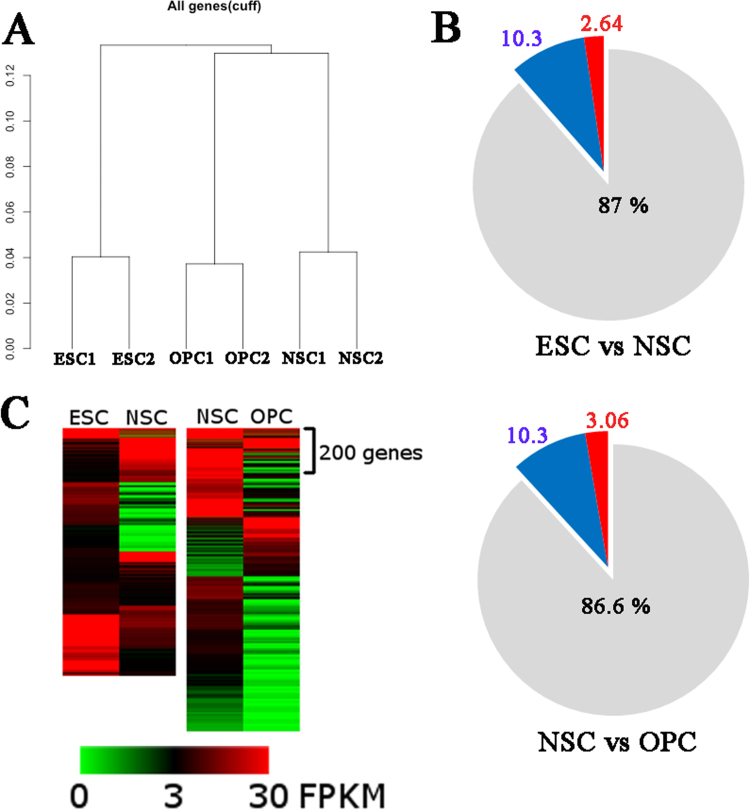
Fig. 2.
Gene expression of human ESCs, NSCs and OPCs. (A) Hierarchical clustering of ESCs, NSCs and OPCs. The pie charts showed percentage of differentially expressed genes of comparing either (B) ESCs to NSCs and NSCs to OPCs. Red indicates a greater than 4-fold increase and blue indicates a greater than 4-fold decrease in genes expression; grey indicates a less than 4-fold difference between cell types (C) Heatmap depicted those genes with greater than 4-fold increased and decreased expression. There were a total of genes (1093 and 1388 genes) listed by absolute log2 in ESCs, NSCs and OPCs. Experiments were repeated on 2 separate occasions. Bar graphs represent the relative expression of the given gene in specific cell type; ***p<0.0005.
2.3. RNA preparation and sequencing
RNA was extracted and isolated from ESC, NSCs and OPCs and purified by utilizing Qiagen RNeasy mini kit (Qiagen), according to the manufacturer's protocol. The total RNA quality was measured using an Agilent 2100 bioanalyzer. To prepare cDNA libraries for sequencing, mRNA-Seq kit was used as per manufacturer's recommended protocol (http://mmjggl.caltech.edu/sequencing/mRNA-Seq_SamplePrep_1004898_D.pdf). Each sample was double sequenced by using Genome Analyzer IIx (Illumina Inc) and generated 50±0.03 bp paired end read length.
2.4. Mapping of RNA-sequence and gene enrichment analysis
The extraction of 50-bp length paired-end reads was achieved using CASAVA (Illumina Pipeline v1.38). For each sample, reads with a quality score of ≥Q30 that passed filtering were used to generate a complete FASTQ file, which was then mapped to UCSC hg19 reference using TopHat2 (v2.0.5) [18]. TopHat2 is a splice-aware aligner and aligns RNA sequences to genomes using the bowtie algorithm. The alignment results were then analyzed by Cufflinks suite [19], which assembles the aligned reads into transcripts and evaluates their relative abundance. The expression of each transcript was quantified as the number of reads mapping to a gene divided by the gene length in kilo bases and the total number of mapped reads in millions, which is called fragments per kilobase of exon per million fragments mapped (FPKM). Then, an R package CummeRbund was used for analyzing the Cufflinks output [20]. The two sets analyzed where genes which showed the highest differential expression between NSCs and ESC, and OPCs compared to NSCs.
2.5. Immunochemistry staining
At the time points indicated, cultured cells were fixed with 4% paraformaldehye and blocked in 20% normal goat serum. Cells were incubated with primary antibody [Oct-3/4, PDGFRα and Nanog (Santa Cruz biotechnology, 1:200 dilution), Nestin and NG2 (Millipore, 1:200 dilution), Pax6 (Covance, 1:200 dilution)] at 4 °C for overnight then washed three times with PBS. Cells were subsequently incubated with goat anti-mouse or anti-rabbit Alexafluor-488 or 594 conjugated secondary antibody [(1:1000 dilution), Invitrogen] for 1 h at room temperature and washed five times with PBS. Stained cells were washed once with H2O, air dried and then mounted with Vectashield solution containing DAPI (Vectorlabs). Staining was analyzed by using Nikon Eclipse 2000 inverted fluorescence microscope. Lineage marker expression values were quantified as the percentage of total DAPI+ cells in each culture field of view that expressed the lineage-specific marker of interest. Data are expressed as the mean percent positive cells±SEM from at least 5 random fields of view from at least 3 independent cultures.
2.6. Western blotting
Cell cultures were lysed with RIPA buffer and protein concentrations were quantified by BCA system (Thermo scientific). 30 μg of each lysate was resolved on an 8% tris-glycine SDS-PAGE and transferred to PVDF membrane (Millipore) using standard techniques. Membranes were blocked and then incubated with specific antibodies Oct3/4 (Santa Cruz, 1:500), Nestin (Millipore, 1:1000), Nanog and NG2 (Cell signaling, 1:1000), at 4 °C overnight. GAPDH (Ambion, 1: 10,000) served as a loading control. Specific antibody labeling of protein bands was detected using horseradish peroxidase conjugated secondary antibodies (Invitrogen) and ECL system (Amersham). Statistical significance was determined using two way Anova from Graph pad (GraphPad Software, Inc, La Jolla, CA, USA).
2.7. Real time-PCR (RT-PCR)
RNA was extracted using Qiagen RNeasy mini kit (Qiagen), according to the manufacturer's protocol. cDNA was synthesized by SuperScript III First-Strand Synthesis System (Invitrogen). We performed RT-PCR utilizing SYBR Green master mix (Bio-Rad) using sequence specific primer in table below. Each reaction was conducted triplicates in CFX96 (Bio-rad) and the condition were as followed: 3 min at 95 °C, 39 cycles of 95 °C for 20 s, 60 °C for 25 s, 72 °C for 25 s and the final step at 95 °C for 10 s. The data was normalized against GAPDH expression and analyzed using delta Ct method [21]. The sequence-specific primers used in these analyses, along with the expected product size are given in the table below. Statistical analysis was done with t-test using excel.
| Gene | Sequence | Size |
|---|---|---|
| ID4-F | ACGACTGCTATAGCCGCCTG | 85 bp |
| ID4-R | ACGTGCTGCAGGATCTCCAC | |
| Hes5-F | ATCCTGGAGATGGCTGTCAG | 98 bp |
| Hes5-R | GAGTAGCCTTCGCTGTAGTC | |
| RARβ-F | AGCTGAGTTGGACGATCTCA | 102 bp |
| RARβ-R | CAGCACTGGAATTCGTGGTG | |
| RARγ-F | GGAAGAAGGGTCACCTGAC | 157 bp |
| RARγ-R | CCAGATCCAGCTGCACGC |
3. Results
3.1. Verification of cell populations for RNA-sequencing
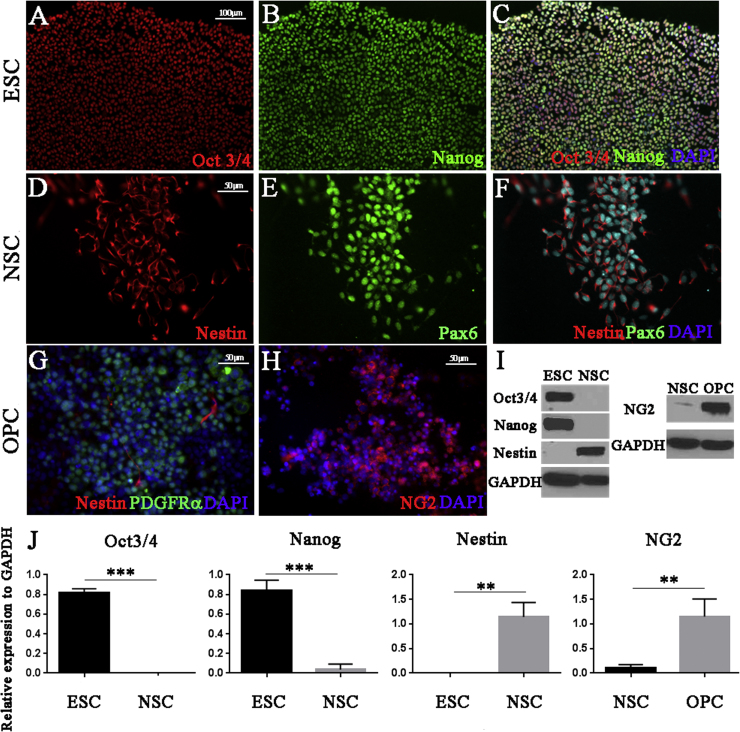
We determined the lineage homogeneity of ESCs, NSCs and OPCs both by immunocytochemistry and western blot analysis. Homogeneous expression of Oct3/4 and Nanog confirmed the ESC lineage (Fig. 1A and B). Similarly, both Nestin and Pax6 were expressed by the great majority (98±3.2%) of cells in the NSCs cultures (Fig. 1D and E). Sorted OPCs, cultured for 24 h following sorting, expressed high levels of the OPC markers PDGFRα and NG2 (Fig. 1G and H) and only a very minor population (2±1%) of this population expressed the NSC marker Nestin (Fig. 1G). We further confirmed the lineage of each of the cell populations by western blot analysis (Fig. 1I). These data show that we could generate highly homogeneous populations of ESCs and NSCs and highly enriched populations of OPCs for this study.
Fig. 1.
Characterization of human ESCs, NSCs and OPCs cell populations. Immunofluorescence staining of specific genes and lineage markers expressed in ESC (A~C), NSC (D–F) and non-sorted OPC (G–H). (A) Oct 3/4 (95±4.3%) and (B) Nanog (89±5.4%) were expressed by all ESC. NSC lineage was confirmed with (D) Nestin (98±2.7%) and (E) Pax6 (87±8.9%) expression. OPC expressed lineage specific markers (G) PDGFRa (65±7.3%) and (H) NG2 (63±7.3%) with less than 2% of cells expressing the immature marker Nestin (H). Cells were counterstained with the nuclear stain DAPI (C, F, G and H). Lineage marker expression values were quantified as the percentage of total DAPI+ cells in each culture field of view that expressed the lineage-specific marker of interest. Data are expressed as the mean percent positive cells±SEM from at least 5 random fields of view from at least 3 independent cultures. Representative western blots of lineage-specific protein expression (I), confirming immunofluorescence staining. (J) The relative expression level was measured against GAPDH. Data is expressed as the mean relative expression of protein±standard deviation of three independent experiments. Analysis of statistical significance was calculated by using t-test from Graph pad. Statistical comparisons of significance are shown with adjoining lines: **p<0.005, ***p<0.0005.
3.2. Differential expression of genes
The transcriptomes of ESCs, NSCs and OPCs were measured by RNA-seq. We obtained up to 43 million paired end reads for which 84–89% of reads aligned to human genome hg19 (Table 1). A comparison of the gene expression by hierarchical clustering showed that a large number of NSC and OPC gene populations were clearly distinguishable from ESCs (Fig. 2A). We compared the differential gene expression between NSCs and OPCs and between ESCs and NSCs. Our data showed that when comparing ESCs to NSCs there were 8440 differentially expressed genes. Of these, 223 genes (2.64%) were more than 4-fold increased in NSCs compared to ESCs whereas 870 genes (10.3%) were more than 4-fold decreased (Fig. 2B). When comparing NSCs to OPCs, there were 9985 differentially expressed genes: 306 genes (3.06%) were up-regulated more than 4-fold in OPCs compared to NSCs, whereas 1032 genes (10.3%) were down-regulated more than 4-fold (Fig. 2B). The variance in expression differed dramatically between the specific lineages of cells and can clearly be seen in the heat map shown in Fig. 2C. Though we have confirmed the lineage of our ESC-derived OPC population by immunohistochemistry (Fig. 1G and H), we also chose to confirm this by analyzing our OPCs for expression of lineage-specific genes. We validated our RNA-seq by assessing those genes that showed the largest difference between ESCs to NSCs, or between OPCs to NSCs (supplementary data 1 and 2 and Supplementary Fig. 1).
Table 1.
Overall information of RNA-sequencing data from ESCs, NSCs and OPC cultures.
| ESC1 | ESC2 | NSC1 | NSC2 | OPC1 | OPC2 | |
|---|---|---|---|---|---|---|
| Total read | 62,164,064 | 61,288,365 | 67,233,095 | 57,918,005 | 59,153,189 | 64,759,513 |
| Paired read | 38,468,038 | 42,060,332 | 43,548,272 | 38,113,156 | 39,940,118 | 43,484,748 |
| % of proper paired read | 61.88 | 63.89 | 61.6 | 62.53 | 63.76 | 63.44 |
| % aligned to human | 88.99 | 88.03 | 87.53 | 85.92 | 84.39 | 86.40 |
3.3. Differential expression of retinoic acid receptor
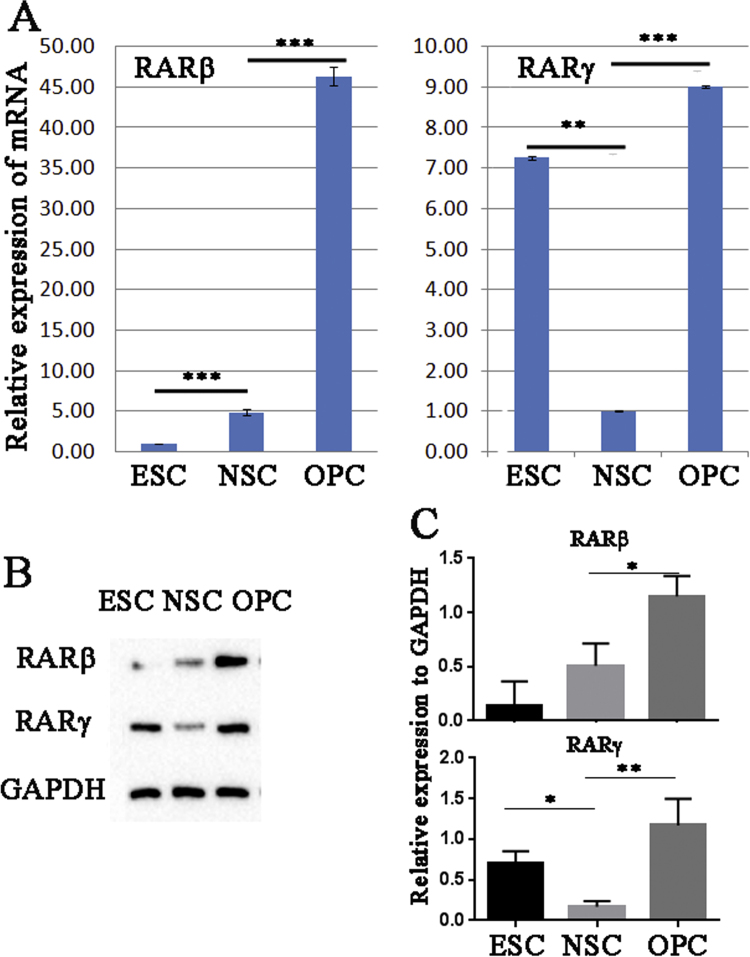
Interestingly, our RNA-seq results pointed to RA signaling as potentially playing an important role following the transition from NSCs to OPCs. The expression of RARβ and RARγ was augmented in OPCs as compared to NSCs, however there was no significant change of RXR (α, β and γ) expression between the two cell types. RARβ showed increased levels of expression at each stage of differentiation from ESCs to NSCs to OPCs (Fragments Per Kilobase of exon per Million fragments mapped (FPKM); 0.17, 0.43 and 7.92; Supplementary Data 1 and 2). Whereas the expression of RARγ was significantly decreased in NSCs as compared to ESCs it increased once again following differentiation to OPC (FPKM; 6.28, 0.50 and 9.53, Supplementary Data 1 and 2). We confirmed these trends using RT-PCR (Fig. 3). The relative expression of RARβ was significantly increased (p<0.0005) in NSCs compared to ESC (4.90±0.31 vs 1.00±0.05) and similarly significantly increased (p<0.0005) in OPCs compared to NSC (46.3±1.11 vs 4.90±0.31). In contrast, RARγ expression was significantly decreased (p<0.005) in NSCs compared to ESCs (1.00±0.01 vs 7.26±0.04) but showed a significant increase (p<0.0005) when NSCs were transitioned to OPCs (1.00±0.01 vs 9.00±0.03).
Fig. 3.
Differential expression of RARβ and RARγ in human ESCs, NSCs and OPCs. Differential gene and protein expression level of RARβ and RARγ was confirmed with RT-PCR (A) and western blotting (B and C). (A) ESCs, NSCs and OPCs were cultured or sorted and RNA was isolated three days after cells were last passaged. (B) Protein was extracted from ESCs, NSCs and OPCs were cultured or sorted. The expression level of RARβ and RARγ was detected by western blotting system. (C) The relative expression level was measured against GAPDH. Data is expressed as the mean relative expression of mRNA or protein±standard deviation of three independent experiments. Analysis of statistical significance was calculated by using two way Anova from Graph pad. Statistical comparisons of significance are shown with adjoining lines: **p<0.005, ***p<0.0005.
3.4. In vitroeffect of RA signaling on transcriptional regulator expression in OPCs
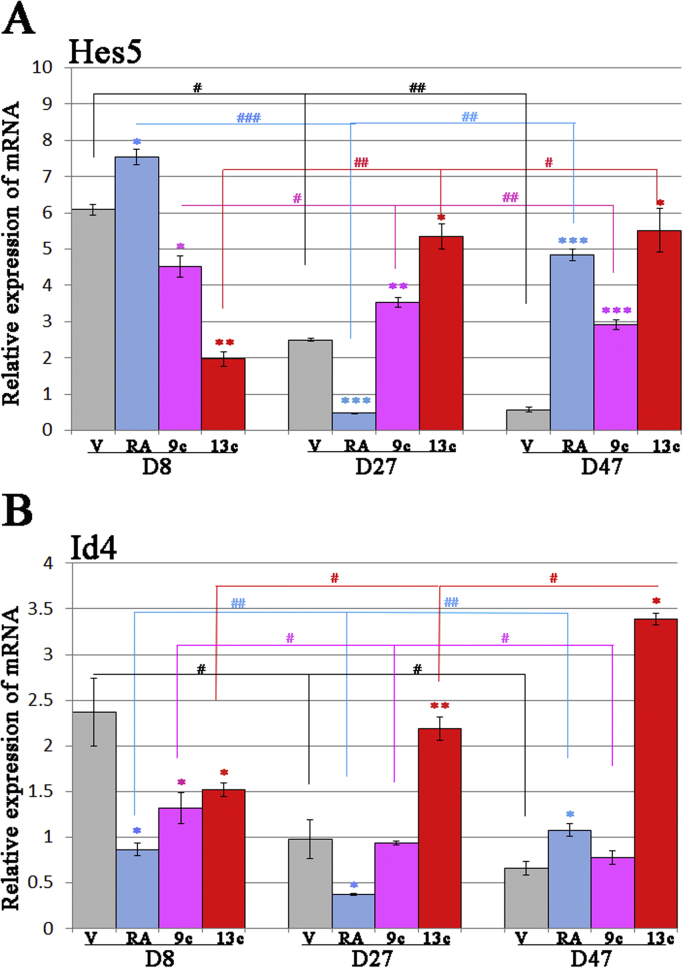
In order to determine if there was any functional significance to the increased expression of RARβ and RARγ in OPCs, we assessed the effects of long-term exposure of hESC-derived OPC to three different isoforms of RA; ATRA, 9cRA or 13cRA. Of these, 9cRA and 13cRA were previously reported to induce differentiation of rodent OPCs into mature oligodendrocytes [10], [22]. To assess the effects of three different RA, RNA was harvested from OPC cultures after 8, 27 and 47 days of exposure, and analyzed using RT-PCR for changes in expression of well-known transcriptional regulators [23]. Our results demonstrated that expression of the negative oligodendrocyte differentiation regulators, Hes5 (hairy and enhancer of split paralogues) and Id4 (inhibitor of DNA binding 4, dominant negative helix-loop-helix protein), was significantly different over time following RA treatment (Fig. 4A and B) whereas other regulators, such as Sox2, Sox6, Oct6, Brn2 and YY1 did not show altered expression (data not shown). In vehicle treated OPC (V: vehicle), both Hes5 (p<0.05) and Id4 (p<0.05) showed the expected significant reduction in expression over time as cells matured. Most strikingly, prolonged (47 days) culture with all three RA's was associated with significantly increased expression of Hes5 by these cells (Fig. 4A). Shorter term (27 days) culture with 9cRA and also caused a significant increase in the expression of Hes5 (P<0.005), in contrast to ATRA which caused a significant reduction at this time point (P<0.0005). The effect of an 8 day exposure was also varied among the RA's: 13cRA and 9cRA were associated with a reduction in Hes5 expression at this stage whereas ATRA was associated with an increase. There was also variant expression of Id4 by OPCs following exposure to the different retinoic acids. All three RA's caused a significant reduction in Id4 expression in the first 8 days. However, prolonged culture with 13cRA for 27 days or 47 days was associated with a significant increase in expression of Id4. There were significant, though less marked, increases in Id4 following culture with ATRA and 9cRA for 47 days. These data when taken together suggest that long-term, sustained treatment with RA may not be conducive to OPC/oligodendrocyte differentiation.
Fig. 4.
Hes5 and Id4 expression following RARβ and RARγ activation in human ESC-derived OPCs. Relative mRNA expression of (A) Hes5 and (B) Id4 in human OPCs following exposure to the three isoforms of RA. OPCs were treated daily with either of 1 μM ATRA, 9cRA or 13cRA. The relative mRNA expression of Hes5 and Id4 was determined by RT-PCR. Data is expressed as the mean relative expression of mRNA±standard deviation of three independent experiments. Analysis of statistical significance was calculated by using two way Anova from Graph pad. Statistical comparisons of significance are shown with adjoining lines: *, ** or *** shows comparisons between each RA treated sample compared to vehicle control at day 8, 27 or 47, respectively. #, ## or ### shows comparisons within each RA treated-sample between day 8 and 27 or day 27 and 47. # and * p<0.05, ## and ** p<0.005, ### and *** p<0.0005.
4. Discussion
The aim of this study was to assess if there are any fundamental gene alterations in RAR gene expression and associated pathways in human OPC, with a view to determining if therapeutic targeting of these pathways could offer any advantage to the neural stem cell niche in a disease such as MS.
Since RNA-seq is emerging as a promising tool in the analysis of gene expression, it is surprising that very few reports was analyzed the gene expression between ESCs and NSCs. To committee neural cell fate from ESC, three signaling pathway, NOTCH, mTOR and TFGb, were involved [24] and expression of Pou3f1 was required [25]. However, no transcriptomic data has specially been updated between OPCs and NSCs. We focused primarily on assessing changes in gene expression during differentiation from OPCs to NSCs. We first confirmed both the homogeneity and lineage of ESCs, NSCs and sorted-OPCs by immunohistochemistry and western blot analysis assessing expression of lineage specific markers (Fig. 1): OPC lineage was also confirmed with the detection of a wide array of OPC specific genes in our RNA-seq data (supplementary data 2). These data are very important for the integrity of this study and not only provide validity to our observations, but enable us to appropriately attribute observed differences to that specific lineage of cells.
Our RNA-seq results demonstrated that very large numbers of genes were alternatively expressed between the 3 different lineages of stem cells (ESCs, NSCs and OPCs). Critically, we observed highly increased expression of RARβ and RARγ in OPCs compared to NSCs, the expression of which was confirmed using RT-PCR and western blotting. Interestingly, the expression of RARβ was gradually increased along with ESCs differentiation to be neural cell fate, NSCs and finally OPCs (Fig. 3). Although the outcome of specific RARβ activation in human CNS was unclear, activation RAR α/β with ATRA might ameliorate remyelination in experimental autoimmune encephalomyelitis mouse for 22 days [26], which was agree to our result that two important transcriptional regulators Hes5 and Id4 were reduced at 27 day after exposure to ATRA. However, longer duration of exposure to ATRA might cause more induction of negative regulators (Fig. 4). Moreover, we determined that exposure of OPC to different RA isoforms ATRA, 9cRA and 13cRA led to significant alterations in the expression of Hes5 and Id4. Compared to control cells the most dramatic effects on Hes5 expression were seen following long-term exposure of OPCs to ATRA and 13cRA whereas the most significant effects on Id4 expression were seen following long-term exposure of OPCs to 13cRA.
Hes5 is one of the targets in Notch signaling along with Hes1, Hesr1 and Hesr2 (HES-related genes) in the nervous system [27]: Hes1, Hesr1 and Hesr2 were not up-regulated genes in our hands. Importantly, it has been shown that the level of Hes5 in oligodendrocyte lineage declined during oligodendrocyte differentiation [28]. Our data suggest that increased Hes5 expression following activation of RAR was related not to Notch signaling but with negative regulation as suggested by Emory [23] who identified Hes5 as an inhibitory regulator during OPCs transition from NSCs. The other regulator, Id4, is also considered one of the essential negative regulators for oligodendrogenesis [29]. Overexpression of Id4 decreased the activity of myelin basic protein (MBP) promoter, which supported its differential role in OPC as a negative regulator [30].
RA is a well-known regulator in neural differentiation and development. In general, this small molecule used one of inducers to generate OPC from NSC [31] and a key regulator to initiate ectoderm fate to be NSC from ESC [15], [16]. However, we concentrated on the role of RA in oligodendrocyte maturation that has been studied in a few publication. Previously, it was shown to inhibit oligodendrocyte maturation during CNS myelination [32], and known as a negative regulator through an RAR-dependent mechanism in the PNS [33], but the down-regulators involved in this inhibitory signaling were not determined. It has been demonstrated that RARα and RARγ were down-regulated in peripheral blood mononuclear cells (PBMC) from MS patients who take dietary vitamin A daily [34]. The analyses of the biology of RA have focused mostly on its interaction with the immune system as shown by the large number of clinical studies using RA as a supplement [35]. It is known fact that RA improves the inflammatory profile of MS patients [36] and a comprehensive review suggested that RA could serve as a potential disease modulator in MS [37]. In contrast, others have described that taking supplementary RA showed no difference to controls when measuring both biochemical parameters [38] and assessing myelin oligodendrocyte glycoprotein (MOG)-reactive T-cells [39]. To date, it has been controversial and no neurological parameters have been reported to be changed by clinically using RA.
In summary, the data we present here have further defined the NSCs to OPCs transcriptome by RNA–seq, describing the upregulation of RAR when NSCs become OPCs. We also show that long-term exposure of OPC to different RA isoforms (ATRA, 9cRA and 13cRA) caused increased expression of inhibitors of OPC differentiation. This may impact the differentiation of OPCs into functional myelinating oligodendrocytes and would suggest that further study into the ubiquitous use of RA in clinical settings is warranted.
Funding
This work was supported by the Race to Erase MS and Department of Neurology Keck School of Medicine Gift Accounts.
Acknowledgements
We give thanks to Arpana Arjun, Maria Ramirez and Jing Du for their technical assistance. We also thank to Dr. Wendy Gilmore for her helpful discussions.
Footnotes
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2016.12.004i.
Contributor Information
Sun young Kim, Email: sunyoungkim0801@gmail.com.
Leslie P. Weiner, Email: lweiner@usc.edu.
Appendix A. Transparency document
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Hu J.G., Fu S.L., Zhang K.H. Differential gene expression in neural stem cells and oligodendrocyte precursor cells: a cDNA microarray analysis. J. Neurosci. Res. 2004;78:637–646. doi: 10.1002/jnr.20317. [DOI] [PubMed] [Google Scholar]
- 2.Dawson M.R., Polito A., Levine J.M., Reynolds R. NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol. Cell. Neurosci. 2003;24:476–488. doi: 10.1016/s1044-7431(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 3.Gensert J.M., Goldman J.E. Endogenous progenitors remyelinate demyelinated axons in the adult CNS. Neuron. 1997;19:197–203. doi: 10.1016/s0896-6273(00)80359-1. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann F., Meinl E. B cells in multiple sclerosis: good or bad guys? An article for 28 may 2014 - World MS Day 2014. Eur. J. Immunol. 2014;44:1247–1250. doi: 10.1002/eji.201470045. [DOI] [PubMed] [Google Scholar]
- 5.Kotter M.R., Stadelmann C., Hartung H.P. Enhancing remyelination in disease--can we wrap it up? Brain. 2011;134:1882–1900. doi: 10.1093/brain/awr014. [DOI] [PubMed] [Google Scholar]
- 6.Miron V.E., Kuhlmann T., Antel J.P. Cells of the oligodendroglial lineage, myelination, and remyelination. Biochim. Biophys. Acta. 2011;1812:184–193. doi: 10.1016/j.bbadis.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Franklin R.J. Why does remyelination fail in multiple sclerosis? Nat. Rev. Neurosci. 2002;3:705–714. doi: 10.1038/nrn917. [DOI] [PubMed] [Google Scholar]
- 8.Mason J.L., Toews A., Hostettler J.D. Oligodendrocytes and progenitors become progressively depleted within chronically demyelinated lesions. Am. J. Pathol. 2004;164:1673–1682. doi: 10.1016/S0002-9440(10)63726-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martino G., Franklin R.J., Baron Van Evercooren A., Kerr D.A. Stem cell transplantation in multiple sclerosis: current status and future prospects. Stem Cells in Multiple Sclerosis (STEMS) Consensus Group. Nat. Rev. Neurol. 2010;6:247–255. doi: 10.1038/nrneurol.2010.35. [DOI] [PubMed] [Google Scholar]
- 10.Huang J.K., Jarjour A.A., Nait Oumesmar B. Retinoid X receptor gamma signaling accelerates CNS remyelination. Nat. Neurosci. 2011;14:45–53. doi: 10.1038/nn.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mora J.R., Iwata M., von Andrian U.H. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat. Rev. Immunol. 2008;8:685–688. doi: 10.1038/nri2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shearer K.D., Stoney P.N., Morgan P.J., McCaffery P.J. A vitamin for the brain. Trends Neurosci. 2012;35:733–741. doi: 10.1016/j.tins.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Gudas L.J., Wagner J.A. Retinoids regulate stem cell differentiation. J. Cell. Physiol. 2011;226:322–330. doi: 10.1002/jcp.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S.C., Wernig M., Duncan I.D., Brüstle O., Thomson J.A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 15.Hu B.Y., Du Z.W., Zhang S.C. Differentiation of human oligodendrocytes from pluripotent stem cells. Nat. Protoc. 2009;4:1614–1622. doi: 10.1038/nprot.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelland E.E., Gilmore W., Weiner L.P., Lund B.T. The dual role of CXCL8 in human CNS stem cell function: multipotent neural stem cell death and oligodendrocyte progenitor cell chemotaxis. Glia. 2011;59:1864–1878. doi: 10.1002/glia.21230. [DOI] [PubMed] [Google Scholar]
- 17.Sim F.J., McClain C.R., Schanz S.J. CD140a identifies a population of highly myelinogenic, migration-competent and efficiently engrafting human oligodendrocyte progenitor cells. Nat. Biotechnol. 2011;29:934–941. doi: 10.1038/nbt.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D., Pertea G., Trapnell C. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C., Hendrickson D.G., Sauvageau M. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C., Robert A., Goff L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 22.Joubert L., Foucault I., Sagot Y. Chemical inducers and transcriptional markers of oligodendrocyte differentiation. J. Neuroci. Res. 2010;88:2546–2557. doi: 10.1002/jnr.22434. [DOI] [PubMed] [Google Scholar]
- 23.Emory B. Regulation of oligodendrocyte differentiation and myelination. Science. 2010;330:779–782. doi: 10.1126/science.1190927. [DOI] [PubMed] [Google Scholar]
- 24.Fathi A., Hatami M., Hajihosseini V. Comprehensive gene expression analysis of human embryonic stem cells during differentiation into neural cells. PLoS One. 2011:e22856. doi: 10.1371/journal.pone.0022856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Q., Song L., Peng G. The transcription factor Pou3f1 promotes neural fate commitment via activation of neural lineage genes and inhibition of external signaling pathways. Elife. 2014:e02224. doi: 10.7554/eLife.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keino H., Watanabe T., Sato Y., Shudo K. Retinoic acid receptor stimulation ameliorates experimental autoimmune optic neuritis. Clin. Exp. Ophthalmol. 2015:558–567. doi: 10.1111/ceo.12308. [DOI] [PubMed] [Google Scholar]
- 27.Louvi A., Artavanis-Tsakonas S. Notch signaling in vertebrate neural development. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 28.Kondo T., Raff M. Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development. 2000;127:2989–2998. doi: 10.1242/dev.127.14.2989. [DOI] [PubMed] [Google Scholar]
- 29.Chen X.S., Zhang Y.H., Cai Q.Y., Yao Z.X. ID2: a negative transcription factor regulating oligodendroglia differentiation. J. Neurosci. Res. 2012;90:925–932. doi: 10.1002/jnr.22826. [DOI] [PubMed] [Google Scholar]
- 30.Marin-Husstege M., He Y., Li J. Multiple roles of Id4 in developmental myelination: predicted outcomes and unexpected findings. Glia. 2006;54:285–296. doi: 10.1002/glia.20385. [DOI] [PubMed] [Google Scholar]
- 31.Park J.C., Jeong W.J., Kim M.Y. Retinoic-acid-mediated HRas stabilization induces neuronal differentiation of neural stem cells during brain development. J. Cell Sci. 2016;129:2997–3007. doi: 10.1242/jcs.184366. [DOI] [PubMed] [Google Scholar]
- 32.Barres B.A., Lazar M.A., Raff M.C. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994;120:1097–1108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- 33.Latasa M.J., Ituero M., Moran-Gonzalez A., Aranda A., Cosgaya J.M. Retinoic acid regulates myelin formation in the peripheral nervous system. Glia. 2010;58:1451–1464. doi: 10.1002/glia.21020. [DOI] [PubMed] [Google Scholar]
- 34.Bitarafan S., Harirchian M.H., Sahraian M.A. Impact of vitamin A supplementation on RAR gene expression in multiple sclerosis patients. J. Mol. Neurosci. 2013;51:478–484. doi: 10.1007/s12031-013-0090-9. [DOI] [PubMed] [Google Scholar]
- 35.Qu Z.X., Pliskin N., Jensen M.W., White D., Arnason B.G. Etretinate augments interferon beta-1b effects on suppressor cells in multiple sclerosis. Arch. Neurol. 2001;58:87–90. doi: 10.1001/archneur.58.1.87. [DOI] [PubMed] [Google Scholar]
- 36.Harrirchian M.H., Mohommadzadeh Honarvar N., Koohdani F. The effect of vitamin a supplementation on disease progression, cytokine levels and gene expression in multiple sclerotic patients: study protocol for a randomized controlled trial. Acta Med. Iran. 2014;52:94–100. [PubMed] [Google Scholar]
- 37.Torkildsen Ø., Løken-Amsrud K.I., Wergeland S., Myhr K.M., Holmøy T. Fat-soluble vitamins as disease modulators in multiple sclerosis. Acta Neurol. Scand. Suppl. 2013;196:16–23. doi: 10.1111/ane.12045. [DOI] [PubMed] [Google Scholar]
- 38.Jafarirad S., Siassi F., Farirchian M.H. The effect of vitamin a supplementation on biochemical parameters in multiple sclerosis patients. Iran. Red Crescent Med. J. 2013;15:194–198. doi: 10.5812/ircmj.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jafarirad S., Siassi F., Harirchian M.H. The effect of vitamin A supplementation on stimulated T-cell proliferation with myelin oligodendrocyte glycoprotein in patients with multiple sclerosis. J. Neurosci. Rural Pract. 2012;3:294–298. doi: 10.4103/0976-3147.102609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material