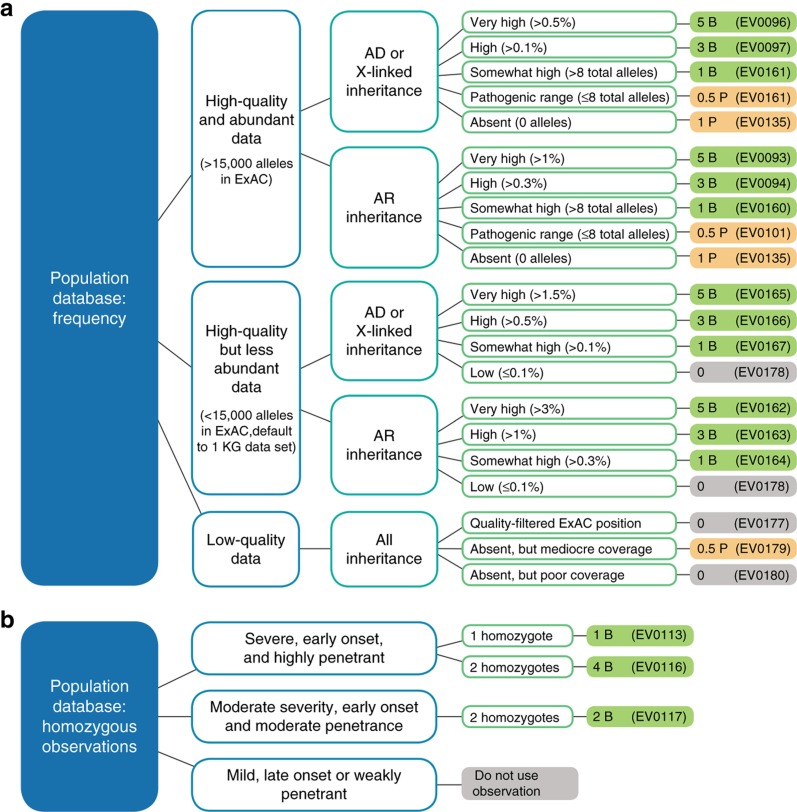
Figure 2.
Population data: Sherloc criteria and decision tree. (a) A single evidence type criterion from the frequency set of criteria is chosen for each variant. This decision tree guides users to the correct criterion based on the quality and abundance of the Exome Aggregation Consortium (ExAC) data at the locus in question, the mode of inheritance of the gene, and the frequency of the variant in ExAC. Points and directionality (pathogenic versus benign) are indicated in the far right column. (b) Decision tree for using observations of homozygotes in the ExAC database depending on the severity, onset, and penetrance of the biallelic phenotype, and the number of homozygotes present. Loci flagged with data quality issues are excluded. Solid orange color corresponds to pathogenic evidence, solid green corresponds to benign evidence, and solid grey corresponds to neutrally weighted evidence. AD, autosomal dominant; AR, autosomal recessive.