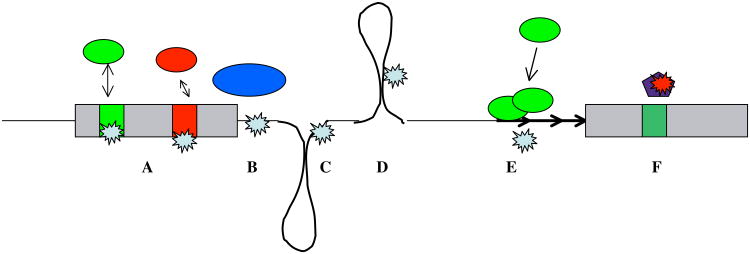
Figure 3. Disease causing mutations.
Exons and introns are indicated as in Figure 2. Arrows indicate repetitive elements. A: Mutation in silencer or enhancer sequences change the interaction between regulatory proteins and pre-mRNA, leading to changes in exon recognition. B: Mutations in splice sites change the interaction with core spliceosomal proteins. C: Due to formation of secondary structures, intronic sequences can be brought close to splice sites, accounting for the effect of some intronic mutations. D: Secondary pre-mRNA structures contribute to exon recognition and their mutations can cause aberrant splicing. E. Expanded repeat elements can sequester splicing regulatory proteins, changing splice site selection in other messages. F. Some snoRNAs regulate splice site selection and their loss can cause disease.