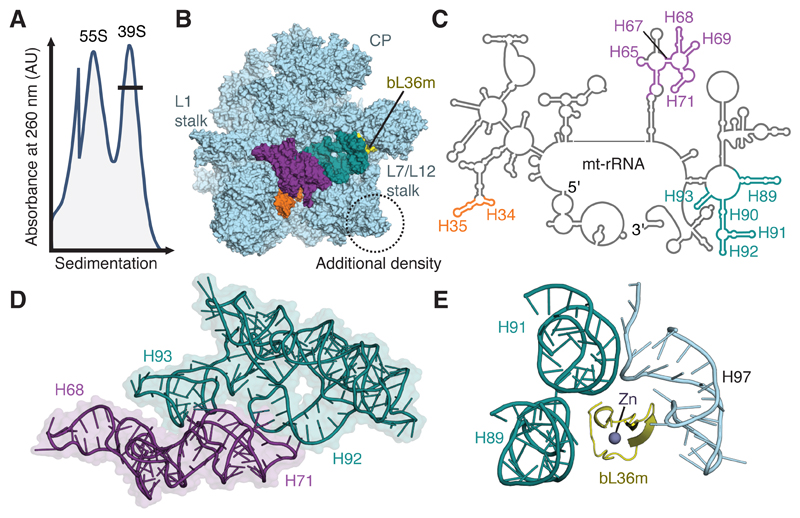
Figure 1. Purification and structural characterization of native mitoribosomal assembly intermediates.
(A) Differential centrifugation separates intact mitoribosomes (55S) from a pool of mt-LSU-like complexes (39S). This pool contains two well-defined assembly intermediates that differ in the presence of folded interfacial rRNA. (B) View of the assembly intermediate with folded interfacial rRNA viewed from the intersubunit interface. Both intermediates feature additional density (circled) relative to the mt-LSU of intact 55S mitoribosomes. However, one class displays unfolded interfacial rRNA with density for H34-35 (orange), H65 and H67-71 (purple), and H89-93 (teal) absent, along with protein bL36m (yellow). Landmark features of the mitoribosome are labeled, including the two stalks and central protuberance (CP). (C) Secondary structure diagram for mt-rRNA with sections of unfolded rRNA colored according to B. (D) Interconnectivity of mt-rRNA helices H67-71 (purple) and H89-93 (teal). (E) In the mature mitoribosome, bL36m coordinates H89 and H91 that are absent in the reconstruction with H97.