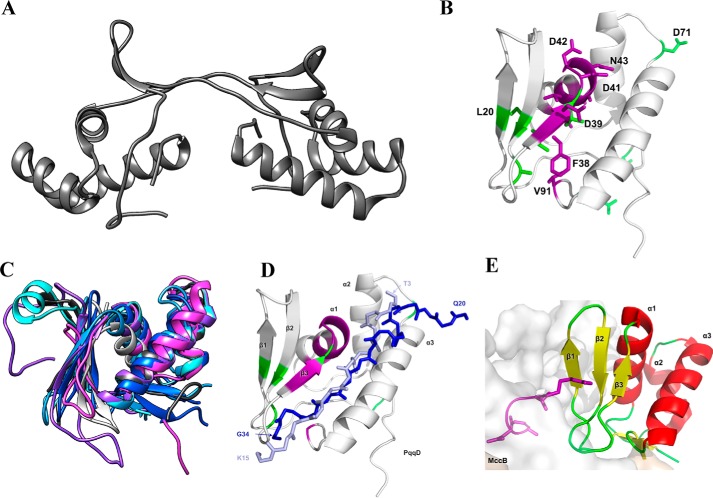
Figure 3.
A, X. campestris PqqD crystal structure (PDB code 3G2B) determined the oligomeric state to be a dimer. B, solution NMR 1H,15N-HSQC experiments on the monomeric M. extorquens AM1 PqqD (PDB code 5SXY) identify residues with significant chemical shifts when bound to PqqA (violet residues) and PqqE (green residues). C, homology-based modeling predicts that the putative peptide chaperone domains of MftB (light blue), QhpD (blue), AlbA (cyan), ThnB (violet), StrB (partial, light gray), and SCIFF maturase (magenta) all have similar structures to that of PqqD (dark gray). D, overlay of the pathway peptides from NisB (light blue) and LynD (dark blue) onto the structure for PqqD, where purple represents residues that are altered in the PqqD–PqqA complex, and green represents positions altered in the ternary complex consisting of PqqD, PqqA, and PqqE. E, complex of MccB with its cognate substrate where the peptide is colored purple.