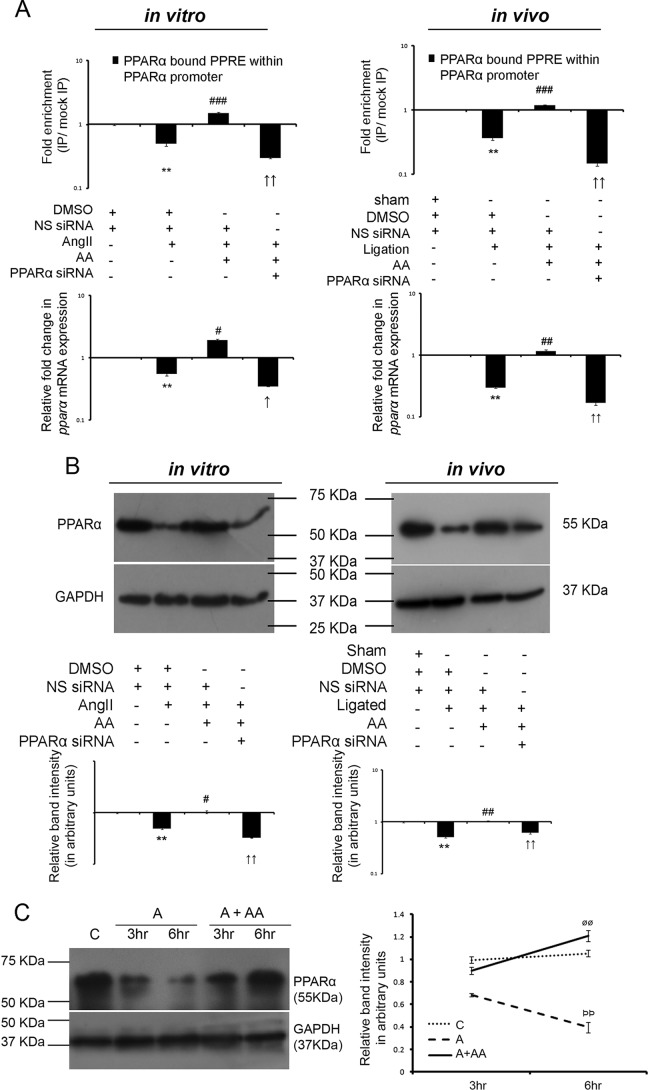
Figure 3.
AA treatment during hypertrophy increases PPARα expression in an autoregulatory loop. A, ChIP assay with anti-PPARα antibody followed by qRT-PCR analyses of the PPRE showing relative binding of PPARα to the PPRE within PPARα promoter among different experimental groups both in vitro and in vivo are represented graphically on logarithmic scale. Hypertrophy samples showed significantly down-regulated-fold enrichment in binding of PPARα to PPRE within the PPARα promoter compared with respective controls. AA treatment in hypertrophy samples further showed significantly increased-fold enrichment of the same, compared with hypertrophy samples. PPARα siRNA treatment in AA-treated hypertrophy groups showing down-regulated-fold enrichment in binding of PPARα to the PPRE compared with the AA-treated hypertrophy samples were used as negative controls. Control and hypertrophy samples in vitro and in vivo were treated with equivalent amounts of DMSO and non-specific (NS) siRNA. AA-treated hypertrophy samples were treated with equivalent amounts of NS siRNA. Chromatin from each experimental group immunoprecipitated with anti-IgG antibody were used for normalization. n = 3 both in vitro and in vivo. Results were analyzed by one-way ANOVA followed by Tukey's post hoc test and expressed as ±S.E. of three independent experiments. **, p < 0.01 with respect to control samples; ###, p < 0.001 with respect to hypertrophy samples; ↑↑, p < 0.01 with respect to AA-treated hypertrophy samples in vitro and/or in vivo. Corresponding changes in pparα mRNA expressions between different experimental groups as observed by qRT-PCR are represented graphically on logarithmic scale. Rpl-32 was used as internal loading control. n = 3 both in vitro and in vivo. Results were analyzed by ANOVA followed by Tukey's post hoc test and expressed as ±S.E. of three independent experiments. **, p < 0.01 with respect to control samples; #, p < 0.05 with respect to hypertrophy samples; ##, p < 0.01 with respect to hypertrophy samples; ↑, p < 0.05 with respect to AA-treated hypertrophy samples; ↑↑, p < 0.01 with respect to AA-treated hypertrophy samples in vitro and/or in vivo. B, Western blot analyses revealed significant decrease in PPARα protein expression during hypertrophy compared with respective control groups that again showed significant recovery in AA-treated hypertrophy samples compared with respective hypertrophy groups in vitro and in vivo. Successful knockdown of PPARα protein expression was also confirmed by PPARα siRNA pretreatment in AA-treated hypertrophy samples. GAPDH was used as internal loading control. Control and hypertrophy samples were treated with equivalent amounts of DMSO and NS siRNA. AA-treated hypertrophy samples were also treated with NS siRNA. Results were analyzed by ANOVA followed by Tukey's post hoc test and expressed as ±S.E. of three independent experiments. n = 10 in vitro, n = 7 in vivo for each experimental group. Representative graphs showing relative changes in expression of PPARα among different experimental groups on logarithmic scale. **, p < 0.01 with respect to control samples; #, p < 0.05 with respect to hypertrophy samples; ##, p < 0.01 with respect to hypertrophy samples; ↑↑, p < 0.01 with respect to AA-treated hypertrophy samples in vitro and/or in vivo. C, Western blot analyses showing time-dependent increase in PPARα expression in AA-treated hypertrophied fibroblasts compared with AngII-treated cells at respective time points under study. GAPDH was used as internal loading control. Results were analyzed by ANOVA followed by Tukey's post hoc test and expressed as ±S.E. of three independent experiments. n = 5 for each experimental group. Representative graphs showing relative changes in PPARα expressions at respective time points among different groups under study. C, control fibroblasts; A, AngII-treated fibroblasts at different time points; A + AA, AngII-treated fibroblasts at different time points, treated along with AA. C and A cells were also treated with equivalent concentration of DMSO. ρρ, <0.01 with respect to A samples at the 3-h time point; øø, <0.01 with respect to A + AA samples at the 3-h time point.