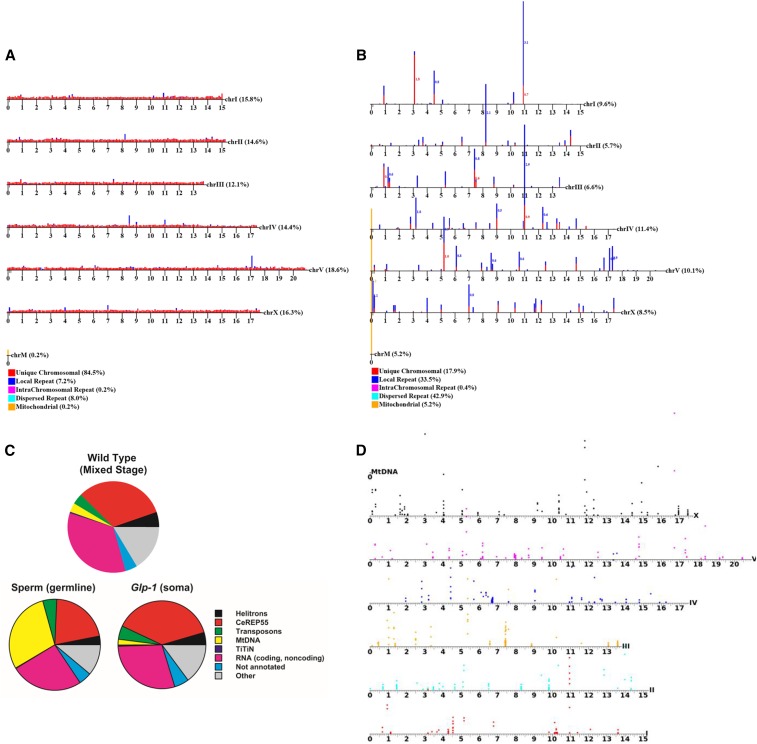
Figure 2.
Data analysis. (A and B) are chromosomal maps of aligned reads in total genomic DNA (G) and eccDNA (Gexo, extrachromosomal circular DNA), respectively. Reads are categorized as: unique, local repeats, intrachromosomal repeats, and dispersed repeats. The graphs show unique reads and focal repeats only, as dispersed repeats cannot be mapped to one location. (C) Whole-genome distribution of sequence classes in eccDNA fractions from WT animals, C. elegans sperm, and animals lacking germline cells. (D) Our methodology applied to glp-1 animals (somatic adults). This map shows uniquely mapped areas on each chromosome that are significantly enriched in the circular pool {1-kbp intervals with enrichment assessed through Bayes maximum-likelihood [minimum of twofold enrichment with a default false discovery rate of 0.05/(2*number of genes)]}. This plot shows only reads that map uniquely to the genome. Position of the colored circle on the y-axis for each interval is proportional to the degree of enrichment.