Abstract
Being an autochthonous species in humans, Lactobacillus gasseri is widely used as a probiotic for fermented products. We thoroughly compared the gene contents of 75 L. gasseri genomes and identified two intraspecific groups by the average nucleotide identity (ANI) threshold of 94%. Group I, with 48 strains, possessed 53 group-specific genes including the gassericin T cluster (9 genes) and N-acyl homoserine lactone lactonase. Group II, with 27 strains, including the type strain ATCC 33323, possessed group-specific genes with plasmid- or phage-related annotations. The genomic differences provide evidences for demarcating a new probiotic group within L. gasseri.
Keywords: comparative genomics, Lactobacillus gasseri, gassericin T, quorum sensing
Lactobacillus gasseri is an autochthonous species of lactic acid bacteria (LAB) that colonizes in the human gastrointestinal tract, vaginal tract, and oral cavity. Its health benefits, such as its antimicrobial activity and probiotic properties, have been well documented [1], making L. gasseri distinct as a probiotic yoghurt inoculum in Japan.
Previously, we reported the existence of two subtypes in L. gasseri by using the average nucleotide identity (ANI), the statistical similarity computed from whole genome sequences [2]. An ANI threshold of 95% corresponds to an experimental DNA-DNA hybridization (DDH) value of 70%, which is the general criterion for a species-level difference [3,4,5]. To reveal genomic characteristics within the two L. gasseri groups, we here report detailed analysis of them focusing on their gene contents.
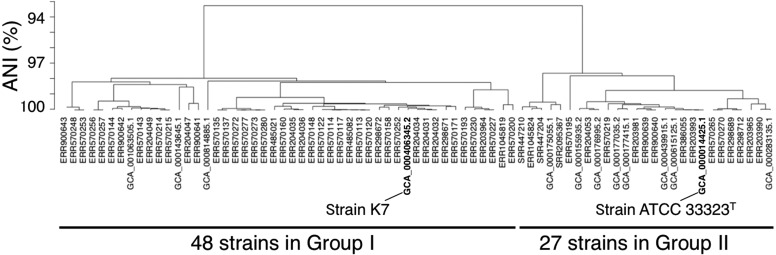
In total, 75 draft genomes of L. gasseri were downloaded from our DFAST Archive of Genome Annotation (DAGA), the curated genome repository of Lactobacillus and Pediococcus from the DDBJ/ENA/GenBank, and Sequence Read Archive (SRA) [2]. They all satisfied a quality rating of ≥4 (out of 5) in our database, meaning that their genome completeness is ≥95% and contamination level is ≤5% as computed by the CheckM software [6]. When ANI values were calculated for all pairs of the obtained genomes by an open-source Python script (https://github.com/widdowquinn/pyani), the 75 genomes were cleanly classified into two groups at the 94% threshold: Group I, consisted of 48 strains, and Group II, consisting of the remaining 27 strains including the type strain (ATCC 33323T, Fig. 1). The genome size ranged from 1.86–2.14 Mb in Group I (average 1.98 Mb) and ranged from 1.78–2.01 Mb in Group II (average 1.89 Mb). The average numbers of protein sequences were 1920 for Group I, 1844 for Group II, and 1893 for both groups (see Supplementary Table for details).
Fig. 1.
The results of hierarchical clustering for 75 L. gasseri strains.
The genome distance is (1-ANI).
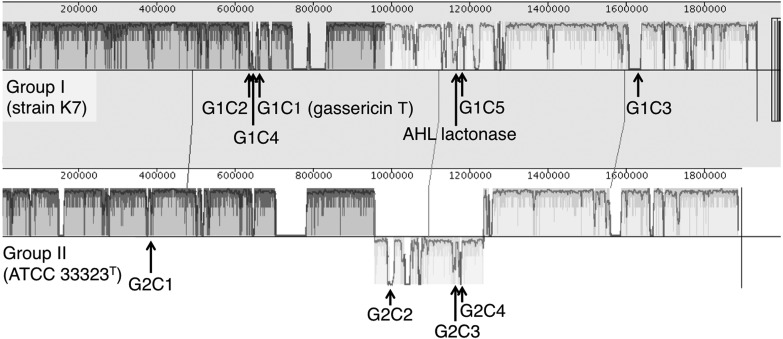
To elucidate the differences between Groups I and II from their gene contents, we computed orthologous gene groups from the overall 141,948 protein sequences using the GET_HOMOLOGUES software (version 1.3) with default settings [7]. Among the 6142 gene groups obtained, 3946 groups contained multiple genes, and 2196 were genome-specific genes, i.e., singletons (Fig. 2). Although the number of genes in each genome was not much different between Group I and Group II, the number of singletons in Group II (27 strains) is far more than in Group I (48 strains). After removing genes of hypothetical/unknown functions, we selected genes whose conservation rates between the two gasseri groups differed by more than 80%. The numbers of Group I- and Group II-specific genes became 53 and 46, respectively. For these genes, we manually verified with the Mauve software whether the selected genes form a gene cluster [8] and identified 5 conserved clusters in Group I and 4 conserved clusters in Group II, respectively (Table 1, 2 and Fig. 3). Notably, we observed a highly conserved gassericin T cluster (cluster ID: G1C1 in Table 1) and a putative N-acyl homoserine lactone (AHL) lactonase in Group I.
Fig. 2.
Venn diagram of the homologous genes in Group I and Group II.
Table 1. Group I-specific orthologous gene groups.
| Cluster ID | Number of genes | Gene names | Conservation (%) |
|
|---|---|---|---|---|
| Group I | Group II | |||
| G1C1 | 10 | lactacin F two-component system inducer peptide precursor (gatP) | 97.9 | 100.0 |
| histidine kinase (gatK) | 95.8 | 3.7 | ||
| two-component system response regulator (gatR) | 95.8 | 0.0 | ||
| peptide ABC transporter ATP-binding protein (gatT) | 95.8 | 3.7 | ||
| lactacin F transporter auxillary protein (gatC) | 95.8 | 3.7 | ||
| bacteriocin (gatA) | 81.3 | 0.0 | ||
| bacteriocin (gatX) | 81.3 | 0.0 | ||
| lactacin F immunity protein (gatI) | 87.5 | 3.7 | ||
| enterocin A immunity protein | 95.8 | 3.7 | ||
| pediocin immunity protein PedB | 97.9 | 96.3 | ||
| G1C2 | 6 | peptidase C45 | 100.0 | 0.0 |
| adenine deaminase | 93.8 | 0.0 | ||
| spermidine/putrescine ABC transporter substrate-binding protein | 93.8 | 0.0 | ||
| spermidine/putrescine ABC transporter ATP-binding protein | 93.8 | 0.0 | ||
| spermidine/putrescine ABC transporter permease protein | 93.8 | 0.0 | ||
| amino acid permease | 100.0 | 0.0 | ||
| G1C3 | 4 | poly(glycerol-phosphate) alpha-glucosyltransferase | 89.6 | 0.0 |
| accessory Sec system protein Asp2 | 87.5 | 0.0 | ||
| preprotein translocase subunit SecA | 87.5 | 7.4 | ||
| preprotein translocase subunit SecY | 83.3 | 0.0 | ||
| G1C4 | 3 | arginine/ornithine antiporter | 87.5 | 0.0 |
| phosphatidylserine decarboxylase | 100.0 | 0.0 | ||
| phosphatidylserine decarboxylase | 87.5 | 0.0 | ||
| G1C5 | 2 | acetolactate synthase large subunit | 100.0 | 3.7 |
| alpha-acetolactate decarboxylase | 100.0 | 3.7 | ||
Gene names follow the output of DAGA annotation, and the cluster ID is our tentative assignment in this Table.
Fig. 3.
Genome alignment by the Mauve software between strain K7 (Group I) and strain ATCC 33323T (Group II).
Positions of group-specific clusters are indicated by Cluster IDs (also see Table 1).
L. gasseri has been reported to produce several bacteriocins, i.e., antimicrobial proteins against its phylogenetic relatives. Gassericin A was reported in L. gasseri LA39 [9], and analogs of gassericin T were reported in L. gasseri SBT2055 (gassericin T) [10], LA158 (gassericin T) [11], LF221 (gassericin K7 B) [12, 13], and EV1461 (gassericin E) [14]. The former, gassericin A, is a rare Class IIc circular bacteriocin. Only the G256_6_33 strain in Group I (SRA: ERR570193) and 4 strains in Group II (G278_2_12, G278_5_18, G287_2_14, and G287_5_2; SRA: ERR570265, ERR570270, ERR900639, and ERR900640, respectively) possessed its close homologs. On the other hand, gassericin T and its analogs are Class IIb two-peptide bacteriocins and have been widely found within L. gasseri. The multiple analogs presumably result from their promiscuous inhibitory spectra that depend on subtle amino acid substitutions or modifications.
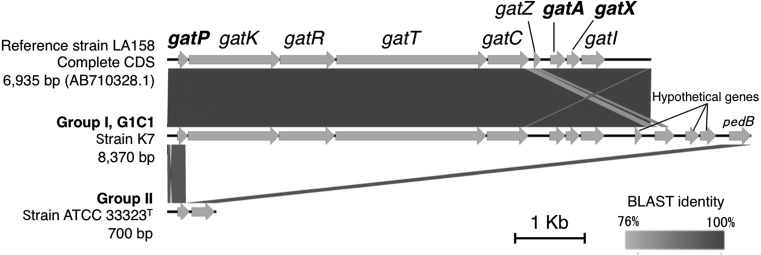
The production of gassericin T requires 9 related genes in a 7 kb genomic region in L. gasseri LA158 (GenBank: AB710328.1) [11]. In Group I, this region was completely conserved in 39 out of 48 strains. Among the remaining 9 strains, seven conserved the partial region (5.2 kb) and lacked the two gassericin T peptide genes (gatA and gatX). Only two out of 48 strains, strain G277_2_5 (SRA: ERR570252) and strain 130918 (GenBank: GCA _000814885.1), lacked the whole region. The regional alignment, created by the Easyfig visualizer (version 2.2.2), is depicted in Fig. 4 [15]. In contrast, all strains in Group II lacked the region except strain G257_1_23 (SRA: ERR570195), which retained a partial region (4.3 kb) without the two peptide genes. In most strains in Group II, however, the two bordering genes, gatP and pedB, were well conserved (Tables 1, 2). GatP is an autoinducer for gassericin T (see later), whereas the function of pedB is has not been determined, and it is hypothetically annotated as “pediocin immunity protein” in the DAGA database.
Fig. 4.
The structure of the gassericin T cluster.
Strain LA158 is used as a reference. See Table 1 for details of the gene functions.
Table 2. Group II-specific orthologous gene groups.
| Cluster ID | Number of genes | Gene names | Conservation (%) |
|
|---|---|---|---|---|
| Group I | Group II | |||
| G2C1 | 3 | death-on-curing family protein | 4.2 | 100.0 |
| NADPH-quinone reductase | 12.5 | 92.6 | ||
| TetR family transcriptional regulator | 2.1 | 100.0 | ||
| G2C2 | 3 | integrase | 0.0 | 96.3 |
| dephospho-CoA kinase | 2.1 | 92.6 | ||
| type III restriction protein, res subunit | 0.0 | 81.5 | ||
| G2C3 | 3 | RelE/StbE family addiction module toxin | 0.0 | 85.2 |
| DNA-damage-inducible protein J | 0.0 | 88.9 | ||
| protein-tyrosine phosphatase | 0.0 | 81.5 | ||
| G2C4 | 2 | flavoprotein | 4.2 | 100.0 |
| LysR family transcriptional regulator | 4.2 | 92.6 | ||
Gene names follow the output of DAGA annotation.
Among the 48 Group I strains, an additionally conserved gene was the AHL lactonase related to quorum sensing. It is a communication system of bacteria to coordinate population. Gram-positive bacteria typically use secreted peptides as autoinducers, and gatP is known as the autoinducer gene for gassericin T production [14]. On the other hand, Gram-negative bacteria often use small molecules such as AHLs and Autoinducer-2 (AI-2 or furanosyl borate diester) [16]. The AHL lactonase is therefore a quorum-quenching enzyme, hydrolyzing the lactone ring of AHLs. As a member of the metallo-hydrolase superfamily [17], this lactonase has been found in various genera such as Bacillus [18], Agrobacterium [19], Rhodococcus [20], and Streptomyces [21].
In Group I, the AHL lactonase was fully conserved in 42 strains, and 4 strains conserved slightly shorter protein sequences. Two strains, G277_2_5 and UMB0099 (SRA: ERR1045819), lacked the gene, and the former strain did not possess the gassericin T cluster either. The amino acid sequence similarity among the 42 Group I strains was 91.1% (256/281 residues). The amino acid similarity between AHL lactonases in L. gasseri K7 and Bacillus sp. 240B1 (GenBank: AF196486.1) was 23.5% (66/281 residues). The gene was not found in Group II.
The extremely high correlation between the gassericin T gene cluster and the AHL lactonase gene in Group I is noteworthy. The gene cluster is chromosomal (at least in complete genomes), showing a good contrast to the gassericin A gene cluster (4 kb) on a plasmid of the producer strain L. gasseri LA39 (JCM11657; the strain was not included in our study due to the absence of the whole genome sequence) [22]. In our study, the gassericin A cluster was found in only one strain in Group I (3 kb partial match in ERR570193) and 4 strains in Group II (a 4 kb complete match in ERR570265 and ERR570270 and a 3.7 kb partial match in ERR900639 and ERR900640). From the draft sequence information, it is hard to tell whether they are plasmidal or chromosomal. However, Group II strains possess more genome-specific genes, and their conserved clusters also contain plasmid- or phage-related annotations such as “TetR transcriptional regulation,” “integrase,” or “RelE/StbE toxin-antitoxin” (Table 2).
These facts together with the ANI analysis demarcate a new probiotic group within L. gasseri. To investigate the functionality of the gassericin T cluster and to biochemically characterize Group I strains, we are conducting an investigation with a polyphasic taxonomic approach (in preparation).
Funding
This study was supported by the Database Integration Coordination Program of the National Bioscience Database Center (Japan), the commission for the Development of Artificial Gene Synthesis Technology for Creating Innovative Biomaterial from the Ministry of Economy, Trade and Industry (METI), Japan, and NIG-JOINT (grant number 2016-53, 2016-54).
Acknowledgments
Computational analysis was performed on the NIG supercomputer at the Research Organization of Information and Systems (ROIS).
References
- 1.Selle K, Klaenhammer TR. 2013. Genomic and phenotypic evidence for probiotic influences of Lactobacillus gasseri on human health. FEMS Microbiol Rev 37: 915–935. [DOI] [PubMed] [Google Scholar]
- 2.Tanizawa Y, Fujisawa T, Kaminuma E, Nakamura Y, Arita M. 2016. DFAST and DAGA: web-based integrated genome annotation tools and resources. Biosci Microbiota Food Health 35: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wayne LG. 1988. International Committee on Systematic Bacteriology: announcement of the report of the ad hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Zentralbl Bakteriol Mikrobiol Hyg [A] 268: 433–434. [DOI] [PubMed] [Google Scholar]
- 4.Stackebrandt E, Ebers J. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33: 152–155. [Google Scholar]
- 5.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79: 7696–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14: 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey N, Malik RK, Kaushik JK, Singroha G. 2013. Gassericin A: a circular bacteriocin produced by lactic acid bacteria Lactobacillus gasseri. World J Microbiol Biotechnol 29: 1977–1987. [DOI] [PubMed] [Google Scholar]
- 10.Kawai Y, Saitoh B, Takahashi O, Kitazawa H, Saito T, Nakajima H, Itoh T. 2000. Primary amino acid and DNA sequences of gassericin T, a lactacin F-family bacteriocin produced by Lactobacillus gasseri SBT2055. Biosci Biotechnol Biochem 64: 2201–2208. [DOI] [PubMed] [Google Scholar]
- 11.Yasuta N, Arakawa K, Kawai Y, Chujo T, Nakamura K, Suzuki H, Ito Y, Nishimura J, Makino Y, Shigenobu S, Saito T. 2014. Genetic and biochemical evidence for gassericin T production from Lactobacillus gasseri LA158. Milk Sci 63: 9–17. [Google Scholar]
- 12.Zorič Peternel M, Čanžek Majhenič A, Holo H, Nes IF, Salehian Z, Berlec A, Rogelj I. 2010. Wide-inhibitory spectra bacteriocins produced by Lactobacillus gasseri K7. Probiotics Antimicrob Proteins 2: 233–240. [DOI] [PubMed] [Google Scholar]
- 13.Mavrič A, Tompa G, Trmčić A, Rogelj I, Bogovič Matijašić B. 2014. Bacteriocins of Lactobacillus gasseri K7—monitoring of gassericin K7 A and B genes’ expression and isolation of an active component. Process Biochem 49: 1251–1259. [Google Scholar]
- 14.Maldonado-Barragán A, Caballero-Guerrero B, Martín V, Ruiz-Barba JL, Rodríguez JM. 2016. Purification and genetic characterization of gassericin E, a novel co-culture inducible bacteriocin from Lactobacillus gasseri EV1461 isolated from the vagina of a healthy woman. BMC Microbiol 16: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27: 1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papenfort K, Bassler BL. 2016. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol 14: 576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong YH, Wang LY, Zhang LH. 2007. Quorum-quenching microbial infections: mechanisms and implications. Philos Trans R Soc Lond B Biol Sci 362: 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong YH, Gusti AR, Zhang Q, Xu JL, Zhang LH. 2002. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl Environ Microbiol 68: 1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlier A, Uroz S, Smadja B, Fray R, Latour X, Dessaux Y, Faure D. 2003. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-Acyl homoserine lactonase activity. Appl Environ Microbiol 69: 4989–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SY, Hwang BJ, Shin MH, Kim JA, Kim HK, Lee JK. 2006. N-acylhomoserine lactonase producing Rhodococcus spp. with different AHL-degrading activities. FEMS Microbiol Lett 261: 102–108. [DOI] [PubMed] [Google Scholar]
- 21.Park SY, Kang HO, Jang HS, Lee JK, Koo BT, Yum DY. 2005. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl Environ Microbiol 71: 2632–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawai Y, Kusnadi J, Kemperman R, Kok J, Ito Y, Endo M, Arakawa K, Uchida H, Nishimura J, Kitazawa H, Saito T. 2009. DNA sequencing and homologous expression of a small peptide conferring immunity to gassericin A, a circular bacteriocin produced by Lactobacillus gasseri LA39. Appl Environ Microbiol 75: 1324–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]