Abstract
Acute hepatopancreatic necrosis disease (AHPND) is a severe shrimp disease originally shown to be caused by virulent strains of Vibrio parahaemolyticus (VPAHPND). Rare cases of AHPND caused by Vibrio species other than V. parahaemolyticus were reported. We compared an AHPND-causing V. campbellii (VCAHPND) and a VPAHPND isolate from the same AHPND-affected pond. Both strains are positive for the virulence genes pirABvp. Immersion challenge test with Litopenaeus vannamei indicated the two strains possessed similar pathogenicity. Complete genome comparison showed that the pirABvp-bearing plasmids in the two strains were highly homologous, and they both shared high homologies with plasmid pVA1, the reported pirABvp-bearing plasmid. Conjugation and DNA-uptake genes were found on the pVA1-type plasmids and the host chromosomes, respectively, which may facilitate the dissemination of pirABvp. Novel variations likely driven by ISVal1 in the genetic contexts of the pirABvp genes were found in the two strains. Moreover, the VCAHPND isolate additionally contains multiple antibiotic resistance genes, which may bring difficulties to control its future outbreak. The dissemination of the pirABvp in non-parahaemolyticus Vibrio also rises the concern of missing detection in industrial settings since the isolation method currently used mainly targeting V. parahaemolyticus. This study provides timely information for better understanding of the causes of AHPND and molecular epidemiology of pirABvp and also appeals for precautions to encounter the dissemination of the hazardous genes.
Keywords: acute hepatopancreatic necrosis disease, Vibrio parahaemolyticus, Vibrio campbellii, plasmid, comparative genomics
Introduction
Acute hepatopancreatic necrosis disease (AHPND) is a severe shrimp disease that has emerged and been causing heavy losses to the global shrimp farming industry since 2010 (Zhang et al., 2012; Tran et al., 2013; Lee et al., 2015). Outbreaks of AHPND have been reported in recent years in Asia (such as China, Vietnam, Malaysia, Philippines, Thailand) and Latin America (such as Mexico) (Gomez-Gil et al., 2014; Gomez-Jimenez et al., 2014; Kondo et al., 2014, 2015; Nunan et al., 2014; Yang et al., 2014; de la Pena et al., 2015; Lee et al., 2015; Soto-Rodriguez et al., 2015; Chonsin et al., 2016; Restrepo et al., 2016; Han et al., 2017). The disease has caused dramatic drops of shrimp production in affected areas, leading to an estimated loss of over $1 billion per year to the global shrimp farming industry (FAO, 2013). Previous studies have revealed that AHPND has a bacterial etiology (Zhang et al., 2012; Tran et al., 2013; Lee et al., 2015). Earlier researches revealed that it is caused by certain virulent strains of Vibrio parahaemolyticus, namely AHPND-causing V. parahaemolyticus (VPAHPND) (Zhang et al., 2012; Tran et al., 2013; Lee et al., 2015). Recently, it was reported that the bacterial etiology of AHPND also includes harveyi-like Vibrio (Kondo et al., 2015), V. owensii (Liu et al., 2015; Xiao et al., 2017) and V. campbellii (Dong et al., 2017a; Han et al., 2017).
V. parahaemolyticus is a Gram-negative bacterium that widely inhabits the marine and estuarine environments and it can cause disease in human and animals (Makino et al., 2003; Thompson et al., 2004). A recent study has demonstrated that VPAHPND harbors a plasmid that expresses a deadly toxin Pirvp (constituted by PirAvp and PirBvp), which is homologous to the Photorhabdus insect-related (Pir) binary toxin (Lee et al., 2015). The Pirvp toxin was firstly characterized in the VPAHPND strain 3HP and was produced by plasmid pVA1 (Lee et al., 2015). The Pirvp toxin is encoded by the pirAvp and pirBvp genes that are located within a fragment delimited by two identical inversely-oriented insertion sequences (IS) ISVal1. It is believed the two ISVal1 with the region in between form a composite transposon called Tn6264 (Han et al., 2017) or pirAB-Tn903 (Xiao et al., 2017). Interestingly, missing of the fragment was seen on a plasmid sharing homology with pVA1 from a non-AHPND-causing strain M2-36, suggesting that a natural deletion or insertion of pirABvp might have occurred (Lee et al., 2015). Other variants with partial deletion were also found (Han et al., 2017). The plasmid pVA1 also carries a cluster of genes related to conjugative transfer (Lee et al., 2015); hence, this plasmid may potentially be able to transfer not only among V. parahaemolyticus strains but also to different bacterial species. Indeed, the existence of pirABvp has been reported in harveyi-like Vibrio (one isolate from Vietnam) (Kondo et al., 2015), V. owensii (one isolate from China) (Liu et al., 2015; Xiao et al., 2017) and V. campbellii (one isolate from China and four from Latin America) (Dong et al., 2017a; Han et al., 2017). We have previously reported the first AHPND-causing V. campbellii (VCAHPND) strain, 20130629003S01, isolated from Guangxi, China, which contains the pirABvp and showed pathogenicity in shrimp (Dong et al., 2017a).
In order to better understand of the spread of pirABvp, in this study, we compared the pathogenicity and genomic features of the previously reported VCAHPND with that of an isolate of VPAHPND from the same batch of shrimp with AHPND in the same pond. Both strains were pirABvp-positive and had a similar pathogenic capacity. We further compared their complete genomes and found that the sequences of the pirABvp-bearing plasmids of both strains were highly homologous, and they shared high homologies with the sequence of pVA1. It suggests that plasmid-mediated interspecies transfer of the hazard genes might have occurred.
Materials and methods
Sample collection, bacteria isolation and identification
In June of 2013, samples were collected from AHPND-suspected shrimp farms in Guangxi, China. Hepatopancreas (HP) from diseased shrimp were aseptically disaggregated and streaked on thiosulfate citrate bile salts sucrose (TCBS) plates at 28°C for 12 h. After pure cultures were obtained, a partial 16S rRNA region was amplified with primers 16S_27F and 16S_1492R (Lane, 1991) (Table S1) and sequenced. Partial rpoD, rctB, and toxR genes were amplified and sequenced as described by Pascual et al. (2010) (Table S1). The concatenated sequences of 16S rRNA, rpoD, rctB, and toxR loci were aligned. Then the phylogenetic tree was constructed using neighbor-joining analysis with maximum composite likelihood model in MEGA 5 (Tempe, AZ, USA) with 1,000 bootstrap replications. The nucleotide sequences from strain Vp 2S01 have been submitted to the GenBank database under accession numbers MF621565 (16S rRNA), MF621566 (toxR), MF621567 (rpoD) and MF621568 (rctB). 16S rRNA, rpoD, rctB and toxR allele sequences of Vc 3S01 were described by our previously report (Dong et al., 2017a). All 16S rRNA, rpoD, rctB, and toxR allele sequences of reference strains were described by Pascual et al. (2010).
Detection of pirAvp and pirBvp in the VPAHPND isolate
Bacterial isolates Vp 2S01 and Vc 3S01 were cultured overnight in Tryptic soy broth with 2% NaCl (TSB+) at 28°C. One milliliter (mL) of the broth culture was boiled for 10 min at 95°C, and the supernatant was obtained by centrifugation, diluted 10-fold with distilled water and used as the template for PCR. PCR was performed using primers VpPirA and VpPirB (Han et al., 2015) (Table S1) as previously described. Protein products were examined by SDS-PAGE and mass spectrometry as previously described (Laemmli, 1970; Wang et al., 2011).
Ethics statement
Since the Ethical Principles and Guidelines for the Use of Animals of the National Research Council of China applies to vertebrates only, there is no official standard for invertebrates, we adapted its principles to shrimp.
Litopenaeus vannamei challenge test
Before the challenge test, ~1 g healthy white shrimp (L. vannamei) were acclimated in the laboratory for 3 days in 50 L seawater at salinity 30 with constant aeration in plastic tanks (density 20 shrimp/tank) at 27 ± 2°C. For the immersion challenge, 20130629002S01 and 20130629003S01 were cultured in TSB+ at 28°C until the OD600 reached 0.8–0.9 (approximately 8–12 h). Immersion challenge was performed following the immersion bioassay protocol described by Tran et al. (2013). Simply, the bacterial suspension was adjusted to 1 × 108 cfu·mL−1 (with TSB+), and 20 shrimp in each group were immersed in 4 L of this suspension for 15 min. The shrimp and bacterial suspension were then poured into tanks containing 50 L of seawater to give a final bacterial density of 1 × 106 cfu·mL−1. The control shrimp were immersed in TSB+ medium. The mortality of each group was recorded every 6 h. Moribund shrimp were fixed with Davidson's alcohol-formalin-acetic acid (DAFA) fixative for histopathological examination. All experiments were done in triplicate.
Histological confirmation of AHPND
All shrimp sampled for histopathology purposes were fixed with DAFA for 24 h and stained with hematoxylin and eosin (H&E) using routine histological methods described by Lightner (1996). The histological sections were analyzed and photographed by a light microscopy system.
Sequencing, assembly and annotation of the genome of the VPAHPND isolate
Genomic DNA isolation, sequencing, assembly and annotation were performed as previously described (Dong et al., 2017b). Antibiotic resistance genes were searched against the ResFinder (Zankari et al., 2012). DNA-uptake genes were identified by Blast against the counterparts reported in Vibrio cholera (Seitz and Blokesch, 2013).
The average nucleotide identity (ANI) of genome sequences of VPAHPND isolates
The complete reference genome sequences of V. parahaemolyticus strains which downloaded from NCBI were used for comparative genome analysis. The reference V. parahaemolyticus strains were M0605 (JALL00000000), D4 (MYFH00000000), FIM-S1708+ (JPLV00000000), NCKU_CV_CHN (JPKU00000000), TUMSAT_DE1_S1 (BAVF00000000), TUMSAT_DE2_S2 (BAVG00000000), NCKU_TV_3HP (JPKS00000000), NCKU_TV_5HP (JPKT00000000), TUMSAT_D06_S3 (BAVH00000000), 1,335 (MYFF00000000), 12297B (MYFG00000000) and A3 (JOKE00000000). ANI value (Richter and Rossello-Mora, 2009) among genomes of Vp 2S01 and reference strains was calculated using the JSpecies program. All strains were cut into fragments of 1,020 bp for calculating the ANI values by using the BLAST algorithm (Goris et al., 2007). Next, a distance dendrogram was constructed using the R program.
Mauve-multiple sequence alignment of pirABvp-bearing plasmids
The complete sequences of pirABvp-bearing plasmids downloaded from NCBI were subject to multiple alignment with pVPGX1 and pVCGX1. Those plasmids were pLA16-2 (accession no. CP021148) harbored by VCAHPND strain LA16-V1, pVHvo (accession no. KX268305) harbored by VOAHPND strain SH14, pVPA3-1 (accession no. KM067908) harbored by VPAHPND strain 13-028A3, pVA1 (accession no. KP324996) harbored by VPAHPND strain 3HP, pVPE61a (accession no. AP014860) harbored by VPAHPND strain VPE61, pV110 (accession no. KY498540) harbored by VPAHPND strain v110. Multiple plasmid sequence alignment was performed using Mauve (Darling et al., 2004).
Antibiotic susceptibility test
Antibiotic susceptibility was determined by the disk diffusion test as described elsewhere (Roque et al., 2001). Briefly, strain suspensions (0.5 MacFarland) of 20130629002S01 and 20130629003S01 were inoculated by lawn onto marine agar and the antimicrobial sensitivity discs positioned. For the present study, antibiotics tested were ampicillin (AMP, 10 μg), aztreonam (ATM, 30 μg), bacitracin (BAC, 0.04 U), cefazolin (CFZ, 30 μg), ceftazidime (CAZ, 30 μg), ceftriaxone (CRO, 30 μg), cephalexin (LEX, 30 μg), cephradine (Rad, 30 μg), ciprofloxacin (CIP, 5 μg), florfenicol (FLO, 30 μg), imipenem (TPM, 10 μg), nitrofurantoin (NIT, 300 μg), norfloxacin (NOR, 10 μg), penicillin (PEN, 10 U), streptomycin (EST, 10 μg), sulfamethoxazole (SMZ, 300 μg), sulfazotrim (SUT, 25 μg), and tetracycline (TCY, 30 μg). The plates were incubated for 24 h at 37°C and the inhibition halos were measured (mm) with vernier caliper. Breakpoints were defined by the guideline (M45-A2, 2010) of Clinical and Laboratory Standards Institute (CLSI). Breakpoints of the antibiotics not listed in the guideline were defined as described by Zhang et al. (2012).
Statistics
All statistical analyses were performed with SPSS, version 17.0 (SPSS Inc., Chicago, IL, USA). Cumulative mortality was compared by using One-Way ANOVA test. A P-value less than 0.05 was considered statistically significant.
Nucleotide sequence accession numbers
Complete genome sequences of 20130629002S01 (V. parahaemolyticus) have been deposited in GenBank under the accession CP020034-CP020037.
Results
Two distinct pirABvp-positive strains isolated from an AHPND epidemic pond displayed similar pathogenicity
In June of 2013, an AHPND-suspected outbreak occurred in a shrimp farm in Guangxi. We have previously reported an AHPND-causing isolate 20130629003S01 (3S01 for short) from the farm, which has been identified as a pirABvp-positive V. campbellii (Dong et al., 2017a). In addition, we isolated another strain, 20130629002S01 (2S01 for short) from the same batch of diseased Litopenaeus vannamei in a same pond of the farm. We found strain 2S01 was a pirABvp-positive V. parahaemolyticus (Figure 1). Thus, for short, we used “Vp 2S01” for strain 2S01 and “Vc 3S01” for strain 3S01 in this paper.
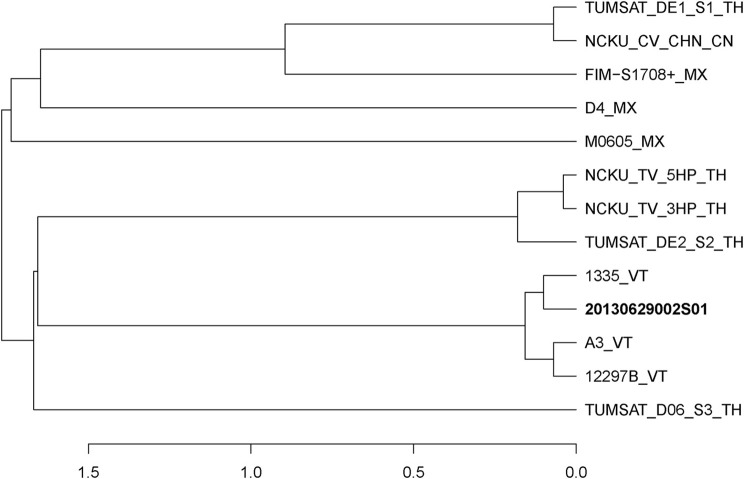
Figure 1.
Distance dendrogram among Vibrio parahaemolyticus strains based on ANI values. The ANI values were calculated using 13 strains, and all values between every two strains were greater than 98%. The complete genome sequences of V. parahaemolyticus (M0605, D4, FIM-S1708+, NCKU_CV_CHN, TUMSAT_DE1_S1, TUMSAT_DE2_S2, NCKU_TV_3HP, NCKU_TV_5HP, TUMSAT_D06_S3, 1335, 12297B and A3) revealed that 20130629002S01 strain has the closest evolutionary relationship (ANI value 99.90%) with an isolate 1,335 isolated from L. vannamei in Viet Nam. VT, Vietnam; TH, Thailand; MX, Mexico; CN, China.
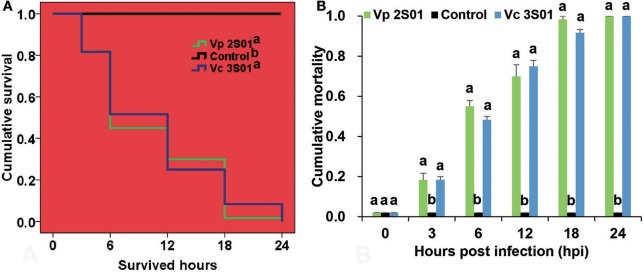
Shrimp immersed with Vp 2S01 or Vc 3S01 suspension were observed to develop typical gross signs of AHPND within 6 h. AHPND shrimp from the Vp 2S01- and Vc 3S01-infected groups had a pale, atrophied HP, and an empty stomach (ST) and midgut (MG), compared to the normal ones that had a normal size HP with dark orange color and a full ST and midgut MG. The survival pattern of different groups was illustrated in Figure 2A. The control group showed no mortality whereas Vp 2S01- and Vc 3S01-infected groups showed 100% mortality within 24 h. As shown in Figure 2B, Vp 2S01 and Vc 3S01 had similar pathogenicity (P > 0.05). Histopathological examination of moribund shrimp samples from Vp 2S01- and Vc 3S01-infected shrimp were used to confirm AHPND, which also revealed similar presence of AHPND lesions in HP (Figure S1).
Figure 2.
Mortalities induced in the immersion bioassay of Litopenaeus vannamei. (A) Survival plots of the shrimp in each group. Survival patterns not sharing a common superscript letter following the pattern curve legends were significantly different from each other (P < 0.05). (B) Mortality of shrimp in each group. Immersion bioassay was performed in triplicate. Error bars indicate standard error of measurement (SEM). Different lowercase letters indicate significant differences (p < 0.05).
Genomic feature comparison of Vp 2S01 and Vc 3S01
The genome of Vc 3S01 has been previously sequenced (Dong et al., 2017b). Genome sequences of Vp 2S01 were determined here using the PacBio RS II sequencing platforms with 189× coverage. Vp 2S01 contains two circular chromosomes (I and II) and two circular plasmids (pVPGX1 and pVPGX2), while Vc 3S01 contains two circular chromosomes (I and II) and four plasmids (pVCGX1 to pVCGX4) (Dong et al., 2017b). Plasmids pVPGX1 and pVCGX1 are homologous to V. paraheamolyticus plasmids pVA1, pVPA3-1 with overall 99% nucleotide identities and partial pFORC4 (accession no. CP009849) with a 94% identity.
Significantly, genes related to DNA transfer were found in both strains. Conjugative transfer were identified on plasmids pVPGX1, pVCGX1, pVCGX3 and pVCGX4 and the chromosome I of Vc 3S01. Plasmids pVPGX1 and pVCGX1 carry tra-trb genes that are highly similar to the counterparts on pVA1 (Lee et al., 2015). The plasmids pVCGX3, pVCGX4 and the chromosome I of Vc 3S01 contain virB/D clusters (Table S2). The ones on pVCGX3 and the chromosome I are near identical, but distinct to the one on pVCGX4. The virB/D genes encoding a type IV secretion system were well known on the Agrobacterium tumefaciens plasmid Ti as being responsible for effector translocation and DNA conjugation (Shirasu et al., 1990; Christie, 2004). In addition, many other homologous virB/D clusters were characterized to be self-transmissible modules of conjugative plasmids and integrative and conjugative elements (ICEs) (Bi et al., 2013). Moreover, intact sets of genes related to DNA uptake were identified on the chromosomes I of both strains (Table 1). Their products display 35.7–95.4% amino acid identities to the counterparts of V. cholerae O1 biovar El Tor str. N16961, which are also encoded by chromosome I. The DNA-uptake genes in V. cholerae N16961 has been proved to be required for efficient natural transformation (Seitz and Blokesch, 2013).
Table 1.
Genes required for efficient natural transformation of Vp 2S01 and Vc 3S01.
| Reference locus_tag* | Gene_name | Vc 3S01 homologs | Vc 3S01_location | Vc 3S01 % identity | Vp 2S01 homologs | Vp 2S01_location | Vp 2S01 % identity |
|---|---|---|---|---|---|---|---|
| VC0462 | pilT | Vc3S01_0475 | chromosome I 549635…550675 | 86.4 | Vp2S01_0459 | chromosome I 507234…508274 | 87.3 |
| VC0543 | recA | Vc3S01_0535 | chromosome I 611769…612812 | 95.1 | Vp2S01_0520 | chromosome I 571980…573023 | 95.4 |
| VC0857 | VC0857 | Vc3S01_2591 | chromosome I 2927019…2927453 | 41.8 | Vp2S01_2489 | chromosome I 2679066…2679497 | 44.5 |
| VC0858 | VC0858 | Vc3S01_2590 | chromosome I 2926915…2926400 | 37.4 | Vp2S01_2488 | chromosome I 2678932…2678447 | 37.8 |
| VC0859 | VC0859 | Vc3S01_2589 | chromosome I 2926390…2925764 | 37.9 | Vp2S01_2487 | chromosome I 2678438…2677812 | 36.1 |
| VC0860 | VC0860 | Vc3S01_2588 | chromosome I 2925764…2924436 | 36.4 | Vp2S01_2486 | chromosome I 2677812…2676484 | 35.7 |
| VC0861 | VC0861 | Vc3S01_2587 | chromosome I 2924446…2924036 | 62.8 | Vp2S01_2485 | chromosome I 2676494…2676084 | 63.5 |
| VC1612 | VC1612 | Vc3S01_1463 | chromosome I 1610447…1610088 | 63.6 | Vp2S01_1405 | chromosome I 1468383…1467658 | 63.4 |
| VC1879 | comEC | Vc3S01_2263 | chromosome I 2554872…2552614 | 41.0 | Vp2S01_2164 | chromosome I 2321516…2319258 | 41.0 |
| VC1917 | ComEA | Vc3S01_2319 | chromosome I 2617558…2617271 | 58.3 | Vp2S01_2235 | chromosome I 2406333…2406040 | 64.1 |
| VC2423 | pilA | Vc3S01_0564 | chromosome I 643331…642915 | 36.2 | Vp2S01_0548 | chromosome I 603064…602636 | 51.3 |
| VC2424 | pilB | Vc3S01_0563 | chromosome I 642915…641230 | 73.7 | Vp2S01_0547 | chromosome I 602636…600951 | 74.0 |
| VC2425 | pilC | Vc3S01_0562 | chromosome I 641195…639972 | 73.8 | Vp2S01_0546 | chromosome I 600926…599703 | 74.1 |
| VC2426 | pilD | Vc3S01_0561 | chromosome I 639899…639030 | 75.0 | Vp2S01_0545 | chromosome I 599638…598769 | 73.3 |
| VC2630 | pilQ | Vc3S01_0320 | chromosome I 365090…366832 | 77.7 | Vp2S01_0314 | chromosome I 345396…347081 | 77.7 |
| VC2631 | pilP | Vc3S01_0319 | chromosome I 364540…365055 | 56.4 | Vp2S01_0313 | chromosome I 344786…345301 | 57.6 |
| VC2632 | pilO | Vc3S01_0318 | chromosome I 363957…364550 | 72.3 | Vp2S01_0312 | chromosome I 344203…344796 | 72.6 |
| VC2633 | pilN | Vc3S01_0317 | chromosome I 363389…363964 | 60.5 | Vp2S01_0311 | chromosome I 343632…344210 | 62.4 |
| VC2634 | pilM | Vc3S01_0316 | chromosome I 362389…363405 | 49.4 | Vp2S01_0310 | chromosome I 342632…343648 | 48.4 |
| VC2719 | comF | Vc3S01_3047 | chromosome I 3434182…3434907 | 46.6 | Vp2S01_2993 | chromosome I 3221591…3222316 | 47.9 |
Reference VC numbers were described by Seitz and Blokesch (2013).
Antibiotic resistance genes were found in both strains, which will be described later. Neither Vp 2S01 nor Vc 3S01 contain genes (scrB) that encode the sucrose hydrolase. Both Vp 2S01 and Vc 3S01 formed green and round colonies on TCBS plate. However, genes encoding the sucrose hydrolase were found in the genome sequence of AHPND-causing strain SH14 of V. owensii (Liu et al., 2015).
Complete plasmid sequences comparison of pirABvp-bearing plasmids
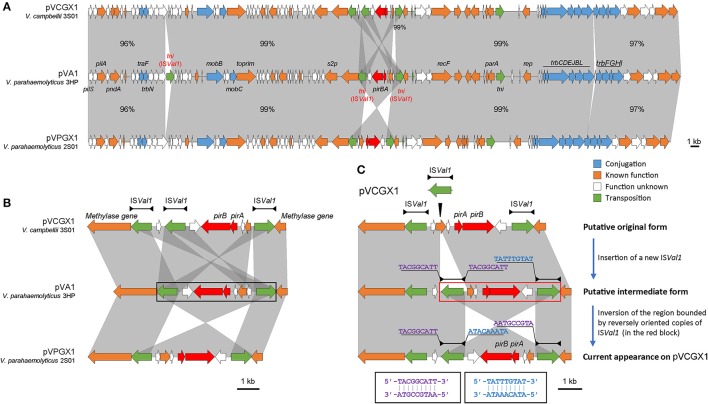
The pirABvp genes located on plasmids pVPGX1 in Vp 2S01 and pVCGX1 in Vc 3S01. The pVPGX1 and pVCGX1 displayed a 99% nucleotide identity spanning the entire sequences, while each of the two plasmids displayed a 96–100% nucleotide identity to pVA1 (Figure 4A). Further comparative analysis found that the two plasmids shared high homologies with other pirABvp-bearing plasmids from AHPND-causing Vibrio strains (Figure S2). Importantly, pVPGX1 and pVCGX1 shared the closest relationship among all the compared plasmids from AHPND-causing Vibrio (Figure 3). More interestingly, different versions of the genetic contexts of pirABvp (pirABvp contexts) were observed in all plasmids when compared (Figure S2).
Figure 3.

Phylogenetic reconstruction based on complete sequences of pirABvp-bearing plasmids. The complete reference sequences of pirABvp-bearing plasmids (pLA16-2, pVHvo, pVPA3-1, pVA1, pVPE61a, pV110) were downloaded from NCBI. Multiple plasmid sequences alignment among pirABvp-bearing plasmids from Vp 2S01, Vc 3S01 and reference strains was calculated using mauve (Darling et al., 2004). VT, Vietnam; TH, Thailand; ED, Ecuador; Mexico; CN, China.
Sequence analysis of the pirABvp contexts of plasmids pVPGX1 and pVCGX1
In pVA1, the pirABvp genes locate within a 5.5-kb fragment ended with two inversely oriented copies of ISVal1 which belonged to the IS903 group of the IS5 family (length includes the two IS elements). The pirABvp were located among some small ORFs between the two ISVal1. ISVal1 was 1,054 bp in length and demarcated by 18-bp perfect inverted repeats (5′-GGCTTTGTTGCGTAATTC-3′). It displayed a 92% nucleotide identity to ISVa2, which was initially found in V. anguillarum (Tolmasky and Crosa, 1995). In pVPGX1, an inversion of the 5.5-kb pirABvp fragment was seen. Transformation of two configurations was likely due to recombination between two opposite ISVal1 (Partridge, 2011). In contrast, the pirABvp contexts in pVCGX1 had a more complex structure. The pirABvp fragment had an extra copy of ISVal1, which divided the fragment into two parts. The left part was inversely syntenic to pVA1, while the right part was directly syntenic to pVA1 (Figure 4B). In addition, an ISVal1 located downstream of trbN and a 217-bp fragment within the trb gene cluster in pVA1 were absent in pVPGX1 and pVCGX1 (Figure 4A).
Figure 4.
Analysis of pirABvp-bearing plasmids pVCGX1 and pVPGX1. (A) Comparison of plasmids pVCGX1 and pVPGX1 with pVA1. Percentages indicate the identities in nucleotide level between syntenic regions. Percentages are not given for 100% identical regions. (B) A zoom-in view of the genetic context of pirABvp. Horizontal lines shown above the schematics, with both ends demarcated by solid triangles to indicated IRs, represent the ISVal1. Black block denotes the 5.5-kb pirABvp fragment describe in the text. (C) Putative evolution process of the genetic context of pirABvp on plasmid pVCGX1. Thin triangle indicates IS insertion. In the red block is the putative region going inversion. Nucleotide sequences flanking the ISVal1 elements are shown in 5′ → 3′. Sequences in identical colors are reverse-complement to each other as denoted in the black blocks. Gene organization is drawn to scale. Red arrows indicate pirABvp genes. Genes with other known functions are denoted using different colors. Syntenic regions were highlighted in gray.
We also analyzed the pirABvp contexts in 12 other sequenced AHPND-causing Vibrio genomes (Table S3). Novel variant was not detected using the current data except that the pirABvp fragment in V. parahaemolyticus M0605 likely located in a different site of a pVA1-type plasmid among them. The site was close to the insertion site of the solitary ISVal1 in pVA1. Note that since most of the reported genomes were unfinished, there is a possibility that there are undetected novel variants lying in the undetermined gaps.
Antibiotic resistance
Antibiotic resistance of Vp 2S01 and Vc 3S01 were detected. The resistance gene profiles of the two strains were also revealed. Vp 2S01 harbored two antibiotic resistance genes tet(35) and tet(34) on chromosome I, while Vc 3S01 contains tet(35) on chromosome I and sul2, strAB, tet(A), strB-2 (extra copy) and floR on pVCGX2. The results of disk diffusion test showed that Vp 2S01 was only susceptible to florfenicol. It was intermediate to ceftriaxone, nitrofurantoin, norfloxacin. Significantly, Vc 3S01 was resistant to a wider spectrum of antibiotics as being resistant or intermediate to all test antibiotics (Table S4).
Discussion
We compared a VPAHPND and a VCAHPND isolate from the same AHPND-affected pond. They displayed a similar pathogenicity and both contained the pirABvp genes carried by plasmids that are highly homologous to pVA1. This study reveals the dissemination of the hazardous genes in V. campbellii, which is likely due to interspecies horizontal gene transfer.
Although VPAHPND is almost the only pathogen known to cause AHPND, studies have indicate that non-parahaemolyticus AHPND-causing Vibrio is emerging. Indeed, certain V. harveyi-like, V. owensii, and V. campbellii strains have been detected to cause AHPND and to be pirABvp-positive (Kondo et al., 2015; Liu et al., 2015; Dong et al., 2017a; Han et al., 2017; Xiao et al., 2017). However, only a limited number of the isolates were characterized and compared. The VCAHPND we previously isolated displays a similar level of pathogenicity to VPAHPND. Immersion challenge for L. vannamei shrimp demonstrates that Vc 3S01-infected shrimp present similar mortality, AHPND manifestations and pathology to Vp 2S01-infected shrimp. It seems that the pathogenicity encoded by pirABvp is independent from host bacteria within closely related Vibrio species. Remarkably, the pirABvp genes may be not originated from V. parahaemolyticus. A recent study has demonstrated that non-virulent V. parahaemolyticus becomes VPAHPND via acquiring a plasmid named pVA1 that expresses a deadly toxin Pirvp (Lee et al., 2015). Additionally, the spread of the VPAHPND via diseased animals and contaminated water accelerates the pandemic of this disease. Similarly, other Vibrio species also could become pathogenic by acquisition of this plasmid. The prevalence of VPAHPND may suggest either they have better capacity of colonization or they are the first pathogenic host bacterium of this plasmid.
The pirABvp-bearing plasmids pVPGX1 and pVCGX1 are highly homologous to pVA1, suggesting the occurrence of horizontal transfer of the pVA1-type plasmid. The pVA1-type plasmids with or without pirABvp genes have been found in many Vibrio species (Xiao et al., 2017). Plasmid is an important factor driving the dynamic gene flow and promoting the fitness of host bacteria (Laurenceau et al., 2013; Seitz and Blokesch, 2013; Matthey and Blokesch, 2016). Plasmid can be acquired mainly through transformation or conjugation. It has been reported that the pVA1 plasmid contained a set of conjugative transfer genes, which suggests that pVA1 might be self-transmissible (Lee et al., 2015). Meanwhile, homologs of genes encoding a DNA-uptake machinery reasonable for natural competence of V. cholera (Seitz and Blokesch, 2013) were also found in the VPAHPND and VCAHPND isolates. Given that the two isolates were from the same origin and the prevalence of VPAHPND, there is a possible scenario that the pVA1-type plasmid pVCGX1 in VCAHPND was originally acquired from VPAHPND, via either self-mediated conjugation or uptake of the free plasmids released by dead VPAHPND cells. Interestingly, the Vc 3S01 contains 4 heterogeneous plasmids. It's unusual that so many different plasmids exist in one Vibrio cells. It may be due to the strain Vc 3S01 has a strong capability to obtain other plasmid. However, whether these transfer genes facilitate the intra-species dissemination of pirABvp still awaits investigation. Moreover, Vc 3S01 is also “armed” with multiple antibiotic resistance genes, which will bring difficulties to control its future spread. Therefore, the transfer of these genes into a new host bacterium not only increases the complexity of causative agents, but also leads to a potential threat with no drug for treatment.
Despite the synteny of the carrier plasmids, the pirABvp genes have dynamic contexts. We assumed the configuration showed in Figure 4C was resulted from the insertion of a new ISVal1 followed by recombination. We subsequently analyzed the flanking sequences of relevant ISVal1 elements to identify the recombination event using the method proposed by Partridge (2011) and found that the right part might have gone through an inversion event after the insertion of a new ISVal1. This right part was bounded by the oppositely oriented middle ISVal1 and right ISVal1. A 9-bp left flank (5′-TACGGCATT-3′) of the middle ISVal1 was reverse-complement to that of the right ISVal1 (5′-AATGCCGTA-3′). If we inverse the current form of this region, the 9-bp direct repeats flanking the middle ISVal1 could be restored and the direction of synteny to pVA1 would be consistent to that of the left part. Thus, an evolution process of pirABvp contexts was deduced. A new ISVal1 inserted into the original pirABvp contexts, generating 9-bp direct repeats (5′-TACGGCATT-3′) and a sub-region that was also bounded by two opposite ISVal1. Then inversion of the sub-region occurred, resulting in the current form.
The pirABvp context initially revealed on pVA1 is bounded by two identical but inversely oriented IS elements, ISVal1. Mobile genetic elements such as IS elements, transposons, integrons and genomic islands are important factors shaping genetic contexts of antimicrobial resistance genes and virulence genes (Partridge, 2011; Stokes and Gillings, 2011). Inversely oriented IS elements close to each other are potential targets for homologous recombination (Partridge, 2011). Unsurprisingly, an inversion form of pirABvp context was found on pVPGX1. It has been suggested that the natural acquisition or deletion of the pirABvp genes is due to transposition or homologous recombination (Lee et al., 2015). However, we prefer the former theory since the recombination results in inversion instead of excision or integration of pirABvp contexts. The two ISVal1 might have formed a composite transposon. Moreover, it seems that ISVal1 is also a factor that complicates the pirABvp context since the more complex configuration on pVCGX1 is very likely due to the insertion of a new ISVal1 followed by recombination. Therefore, we propose that ISVal1 plays a role not just in the translocation of the pirABvp genes but also in the modulation of their contexts.
TCBS plate is currently used as a simple tool in many shrimp farms for monitoring and forewarning the V. parahaemolyticus-associated risk that may cause AHPND, as most V. parahaemolyticus strains develop green colonies (Non-sucrose-fermenting bacterial colonies are covered by the green color). This method works for V. campbellii as well. Indeed, neither Vp 2S01 nor Vc 3S01 contains genes that encode the sucrose hydrolase, such as scrB gene (Reid and Abratt, 2005). Other Vibrio species, like V. alginoluticus, V. cholerae carry the sucrose hydrolase genes and develop yellow colonies. A V. owensii strain from a shrimp farm suffering AHPND in Haiyang of Shandong Province in 2017 was also tested as pirABVP positive. The strain was tested on TCBS plate and developed a yellow colony (unpublished results). Therefore, there is a potential risk of negative detection of pathogenic strains when using the TCBS plate method given the increasing intra-species dissemination of the pirABVP genes. We here suggest both green and yellow colonies should be tested under such circumstance.
AHPND has caused severe production collapses and heavy economic losses in Asia and Latin America. (Zhang et al., 2012; FAO, 2013; De Schryver et al., 2014; Nunan et al., 2014; de la Pena et al., 2015; Lee et al., 2015). Actions to bring the disease to an end are in urgent need. This study demonstrates the dissemination of the pirABvp genes in V. campbellii and thus sheds light on the molecular epidemiology of the virulence genes. Moreover, acquisition of the pVA1-type, pirABvp-bearing plasmids in diverse Vibrio species increases the complexity of causative agents of AHPND and their potential threat to the shrimp industry. Therefore, our study provides timely information for better understanding of the causes, epidemiological features of AHPND, as well as developments of response measures and prevention and control strategies.
Author contributions
XD and DB designed and conducted the study, performed most of the experiments, and as well as HW wrote the manuscript. PZ, QY, XW, MC, CG, and WW performed the biological experiments. ZL assembled preliminary sequences and analysis. HW, QY, GX, XW, YZ, and QW discussed the results and modified the manuscript. JH designed the study and wrote the manuscript. All authors reviewed the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the projects under Central Public-interest Scientific Institution Basal Research Fund, YSFRI, CAFS (20603022016012), the project of the Aoshan Sci. & Tech. Innovation Program of Qingdao National Laboratory for Marine Science and Technology (2015ASKJ02), the China Agriculture Research System (CARS-47), and the Construction Programme for Distinguished Taishan Scholars of Shandong Province of China.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.01859/full#supplementary-material
References
- Bi D., Liu L., Tai C., Deng Z., Rajakumar K., Ou H. Y. (2013). SecReT4: a web-based bacterial type IV secretion system resource. Nucleic Acids Res. 41(Database issue), D660–D665. 10.1093/nar/gks1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chonsin K., Matsuda S., Theethakaew C., Kodama T., Junjhon J., Suzuki Y., et al. (2016). Genetic diversity of Vibrio parahaemolyticus strains isolated from farmed Pacific white shrimp and ambient pond water affected by acute hepatopancreatic necrosis disease outbreak in Thailand. FEMS Microbiol. Lett. 363:fnv222. 10.1093/femsle/fnv222 [DOI] [PubMed] [Google Scholar]
- Christie P. J. (2004). Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim. Biophys. Acta 1694, 219–234. 10.1016/j.bbamcr.2004.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. C., Mau B., Blattner F. R., Perna N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pena L. D., Cabillon N. A., Catedral D. D., Amar E. C., Usero R. C., Monotilla W. D., et al. (2015). Acute hepatopancreatic necrosis disease (AHPND) outbreaks in Penaeus vannamei and P. monodon cultured in the Philippines. Dis. Aquat. Organ. 116, 251–254. 10.3354/dao02919 [DOI] [PubMed] [Google Scholar]
- De Schryver P., Defoirdt T., Sorgeloos P. (2014). Early mortality syndrome outbreaks: a microbial management issue in shrimp farming? PLoS Pathog. 10:e1003919. 10.1371/journal.ppat.1003919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Wang H., Xie G., Zou P., Guo C., Liang Y., et al. (2017a). An isolate of Vibrio campbellii carrying the pirVP gene causes acute hepatopancreatic necrosis disease. Emerg. Microbes Infect. 6, e2. 10.1038/emi.2016.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Wang H., Zou P., Chen J., Liu Z., Wang X., et al. (2017b). Complete genome sequence of Vibrio campbellii strain 20130629003S01 isolated from shrimp with acute hepatopancreatic necrosis disease. Gut. Pathog. 9:31. 10.1186/s13099-017-0180-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2013). Report of the FAO/MARD Technical Workshop on Early Mortality Syndrome (EMS) or Acute Hepatopancreatic Necrosis Syndrome (AHPNS) of Cultured Shrimp (under TCP/VIE/3304). Rep. no. 1053, Hanoi, 25–27. Available online at www.fao.org/docrep/018/i3422e/i3422e.pdf (Accessed July 28, 2015). [Google Scholar]
- Gomez-Gil B., Soto-Rodriguez S., Lozano R., Betancourt-Lozano M. (2014). Draft genome sequence of Vibrio parahaemolyticus strain M0605, which causes severe mortalities of shrimps in Mexico. Genome Announc. 2:e00055-14. 10.1128/genomeA.00055-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Jimenez S., Noriega-Orozco L., Sotelo-Mundo R. R., Cantu-Robles V. A., Cobian-Guemes A. G., Cota-Verdugo R. G., et al. (2014). High-quality draft genomes of two Vibrio parahaemolyticus strains aid in understanding acute hepatopancreatic necrosis disease of cultured shrimps in Mexico. Genome Announc. 2:e00800-14. 10.1128/genomeA.00800-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J., Konstantinidis K. T., Klappenbach J. A., Coenye T., Vandamme P., Tiedje J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57(Pt. 1), 81–91. 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- Han J. E., Tang K. F., Aranguren L. F., Piamsomboon P. (2017). Characterization and pathogenicity of acute hepatopancreatic necrosis disease natural mutants, pirABvp (-) V. parahaemolyticus, and pirABvp (+) V. campbellii strains. Aquaculture 470, 84–90. 10.1016/j.aquaculture.2016.12.022 [DOI] [Google Scholar]
- Han J. E., Tang K. F., Tran L. H., Lightner D. V. (2015). Photorhabdus insect-related (Pir) toxin-like genes in a plasmid of Vibrio parahaemolyticus, the causative agent of acute hepatopancreatic necrosis disease (AHPND) of shrimp. Dis. Aquat. Organ. 113, 33–40. 10.3354/dao02830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H., Tinwongger S., Proespraiwong P., Mavichak R., Unajak S., Nozaki R., et al. (2014). Draft genome sequences of six strains of Vibrio parahaemolyticus isolated from early mortality syndrome/acute hepatopancreatic necrosis disease shrimp in Thailand. Genome Announc. 2:e00221-14. 10.1128/genomeA.00221-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H., Van P. T., Dang L. T., Hirono I. (2015). Draft genome sequence of non-Vibrio parahaemolyticus acute hepatopancreatic necrosis disease strain KC13.17.5, isolated from diseased shrimp in Vietnam. Genome Announc. 3:e00978-15. 10.1128/genomeA.00978-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lane D. J. (1991). Nucleic acid techniques, in Bacterial Systematics, eds Stackebrandt E., Goodfellow M. (New York, NY: Wiley; ), 115–175. [Google Scholar]
- Laurenceau R., Pehau-Arnaudet G., Baconnais S., Gault J., Malosse C., Dujeancourt A., et al. (2013). A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog. 9:e1003473. 10.1371/journal.ppat.1003473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. T., Chen I. T., Yang Y. T., Ko T. P., Huang Y. T., Huang J. Y., et al. (2015). The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. U.S.A 112, 10798–10803. 10.1073/pnas.1503129112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner D. (1996). A Handbook of Shrimp Pathology and Diagnostic Procedures for Diseases of Cultured Penaeid Shrimp. Baton Rouge, LA: World Aquaculture Society. [Google Scholar]
- Liu L., Xiao J., Xia X., Pan Y., Yan S., Wang Y. (2015). Draft genome sequence of Vibrio owensii strain SH-14, which causes shrimp acute hepatopancreatic necrosis disease. Genome Announc. 3:e01395-15. 10.1128/genomeA.01395-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino K., Oshima K., Kurokawa K., Yokoyama K., Uda T., Tagomori K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361, 743–749. 10.1016/S0140-6736(03)12659-1 [DOI] [PubMed] [Google Scholar]
- Matthey N., Blokesch M. (2016). The DNA-uptake process of naturally competent Vibrio cholerae. Trends Microbiol. 24, 98–110. 10.1016/j.tim.2015.10.008 [DOI] [PubMed] [Google Scholar]
- Nunan L., Lightner D., Pantoja C., Gomez-Jimenez S. (2014). Detection of acute hepatopancreatic necrosis disease (AHPND) in Mexico. Dis. Aquat. Organ. 111, 81–86. 10.3354/dao02776 [DOI] [PubMed] [Google Scholar]
- Partridge S. R. (2011). Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 35, 820–855. 10.1111/j.1574-6976.2011.00277.x [DOI] [PubMed] [Google Scholar]
- Pascual J., Macian M. C., Arahal D. R., Garay E., Pujalte M. J. (2010). Multilocus sequence analysis of the central clade of the genus Vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int. J. Syst. Evol. Microbiol. 60(Pt 1), 154–165. 10.1099/ijs.0.010702-0 [DOI] [PubMed] [Google Scholar]
- Reid S. J., Abratt V. R. (2005). Sucrose utilisation in bacteria: genetic organisation and regulation. Appl. Microbiol. Biotechnol. 67, 312–321. 10.1007/s00253-004-1885-y [DOI] [PubMed] [Google Scholar]
- Restrepo L., Bayot B., Betancourt I., Pinzon A. (2016). Draft genome sequence of pathogenic bacteria Vibrio parahaemolyticus strain Ba94C2, associated with acute hepatopancreatic necrosis disease isolate from South America. Genom. Data 9, 143–144. 10.1016/j.gdata.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M., Rossello-Mora R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque A., Molina-Aja A., Bolán-Mejía C., Gomez-Gil B. (2001). In vitro susceptibility to 15 antibiotics of vibrios isolated from penaeid shrimps in Northwestern Mexico. Int. J. Antimicrob. Agents 17, 383–387. 10.1016/S0924-8579(01)00308-9 [DOI] [PubMed] [Google Scholar]
- Seitz P., Blokesch M. (2013). DNA-uptake machinery of naturally competent Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 110, 17987–17992. 10.1073/pnas.1315647110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K., Morel P., Kado C. I. (1990). Characterization of the virB operon of an Agrobacterium tumefaciens Ti plasmid: nucleotide sequence and protein analysis. Mol. Microbiol. 4, 1153–1163. [DOI] [PubMed] [Google Scholar]
- Soto-Rodriguez S. A., Gomez-Gil B., Lozano-Olvera R., Betancourt-Lozano M., Morales-Covarrubias M. S. (2015). Field and experimental evidence of Vibrio parahaemolyticus as the causative agent of acute hepatopancreatic necrosis disease of cultured shrimp (Litopenaeus vannamei) in Northwestern Mexico. Appl. Environ. Microbiol. 81, 1689–1699. 10.1128/AEM.03610-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes H. W., Gillings M. R. (2011). Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into gram-negative pathogens. FEMS Microbiol. Rev. 35, 790–819. 10.1111/j.1574-6976.2011.00273.x [DOI] [PubMed] [Google Scholar]
- Thompson F. L., Iida T., Swings J. (2004). Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68, 403–431. 10.1128/MMBR.68.3.403-431.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky M. E., Crosa J. H. (1995). Iron transport genes of the pJM1-mediated iron uptake system of Vibrio anguillarum are included in a transposonlike structure. Plasmid 33, 180–190. [DOI] [PubMed] [Google Scholar]
- Tran L., Nunan L., Redman R. M., Mohney L. L., Pantoja C. R., Fitzsimmons K., et al. (2013). Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aquat. Organ. 105, 45–55. 10.3354/dao02621 [DOI] [PubMed] [Google Scholar]
- Wang J., Jiang J., Zhang H., Wang J., Cai H., Li C., et al. (2011). Integrated transcriptional and proteomic analysis with in vitro biochemical assay reveal the important role of CYP3A46 in T-2 toxin hydroxylation in porcine primary hepatocytes. Mol. Cell Proteomics 10:M111.008748. 10.1074/mcp.M111.008748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Liu L., Ke Y., Li X., Liu Y., Pan Y., et al. (2017). Shrimp AHPND-causing plasmids encoding the PirAB toxins as mediated by pirAB-Tn903 are prevalent in various Vibrio species. Sci. Rep. 7:42177. 10.1038/srep42177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. T., Chen I. T., Lee C. T., Chen C. Y., Lin S. S., Hor L. I., et al. (2014). Draft genome sequences of four strains of Vibrio parahaemolyticus, three of which cause early mortality syndrome/acute hepatopancreatic necrosis disease in shrimp in China and Thailand. Genome Announc. 2:e00816-14. 10.1128/genomeA.00816-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. C., Liu F., Bian H. H., Liu J., Pan L. Q., Huang J. (2012). Isolation, identification, and pathogenicity analysis of a Vibrio parahaemolyticus strain from Litopenaeus vannamei. Prog. Fishery Sci. (in Chinese). 33, 56–62. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.