Figure 2.
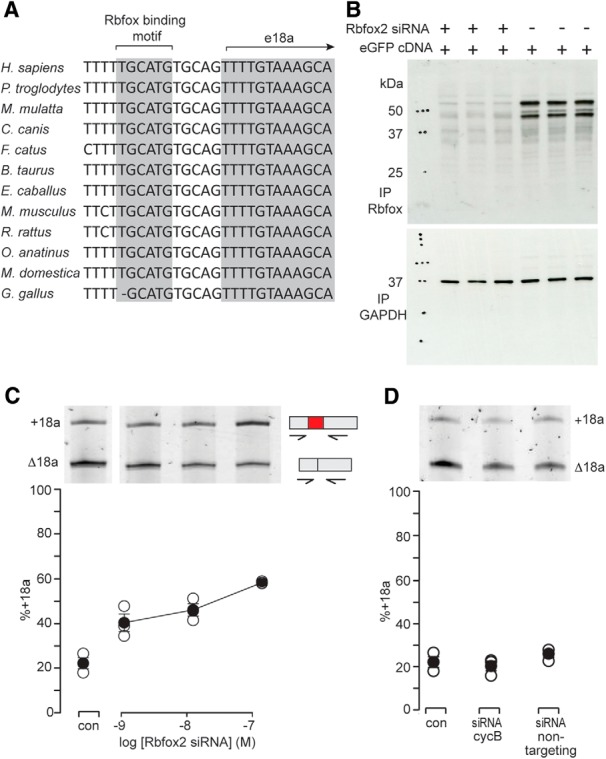
The splicing factor Rbfox2 represses e18a inclusion in a cell line. A, Genome alignments: Rbfox binding motif (U)GCAUG located upstream of e18a (Minovitsky et al., 2005). Putative Rbfox consensus sequence is conserved across all 12 species shown. B, Western blotting shows Rbfox protein in F11 cells transfected with siRNA against Rbfox2 (lanes 1-3) compared to control (lanes 4-6). Anti-Rbfox recognizes RNA binding motif (RRM) of all Rbfox protein isoforms (Gehman et al., 2011). The anti-GAPDH signal from the same membrane stripped and reprobed is shown. The experiment was run in triplicate, lanes 1-3 (siRNA) and lanes 4-6 (control). C, RT-PCR analysis of +18a and Δ18a CaV2.2 mRNAs from F11 cells transfected with Rbfox2-specific siRNA. Three independent transfections were analyzed per condition. E18a RT-PCR signal, as a percentage of total, was: 22.5 ± 2.3% (nontransfected F11) compared to 58.4 ± 2.5% in cells transfected with 100 nM siRNA to Rbfox2 (Student’s unpaired t test, p = 0.000129a). D, RT-PCR analysis of CaV2.2 mRNA extracted from nontransfected F11 cells (con), F11 cells transfected with siRNA to cyclophilin B (cycB siRNA), or transfected with nontargeting siRNA. Representative gel is shown with average % e18a signal in PCR amplification of F11 RNA from three experiments. Average +18a signals were similar: 22.5 ± 2.3% (con); 20.3 ± 2.3% (cycB); and 26.0 ± 1.7% (nontargeting siRNA; one-way ANOVA, F = 1.789, p = 0.246b).
