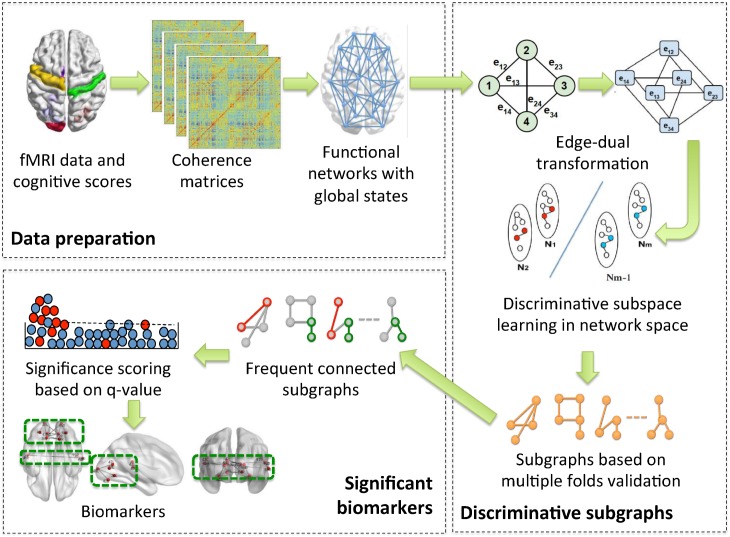
Fig 1. An overview of the method.
We start with experimental fMRI and learning rate measurements from individual sessions. We compute pairwise coherence among all regions and create session-specific functional networks with a global label based on the learning rate. We transform the coherence networks into edge-dual graphs and perform a discriminative subspace learning with multiple-split k-fold validation to produce a set of discriminative connected subgraphs. Next, we mine conserved connected subgraphs identified by subspace learning and compute the significance of their individual accuracy while implementing a false discovery rate correction for multiple comparisons to obtain our final biomarkers.