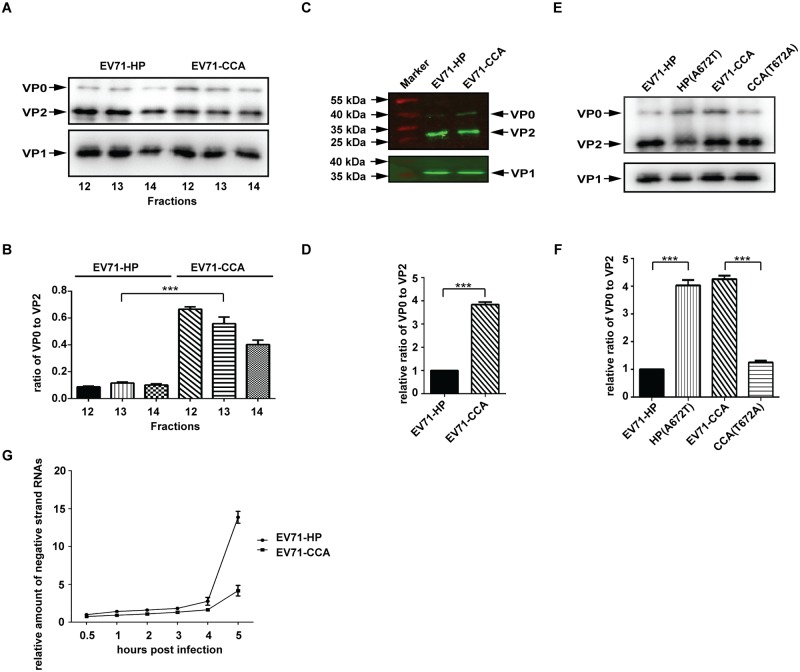
Fig 6. The VP1107 residue regulates the cleavage of precursor VP0.
(A) Protein composition of the 160S particles (fractions of 12–14) of EV71-HP and EV71-CCA. The corresponding fractions of discontinuous sucrose density gradient were concentrated by ultracentrifugation and then subjected to western blotting with anti-EV71 VP0/VP2 and VP1 antibody, respectively. (B) VP0 cleavage efficiency of EV71. The band density of VP0 and VP2 in (A) were quantified using ImageJ, and the data were used to calculate the ratio of VP0 to VP2, which represents the VP0 cleavage efficiency. (C) Western blotting analysis of the 160S particles of EV71 by the Odyssey infrared imaging system. (D) VP0 cleavage efficiency of EV71. The band density of VP0 and VP2 protein shown in (C) was quantified and used to determine VP0 cleavage efficiency. (E) Western blotting analysis of 160S particles of EV71-HP, HP(A672T), CCA and CCA(T672A). (F) VP0 cleavage efficiency of the above substitutional mutants. Three asterisks indicate P<0.01. (G) RNA replication of CCA strain is delayed relative to HP strain. The cells were inoculated with equal amount of HP strain or CCA strain. After synchronized viral adsorption, the infected cells were incubated at 37°C and collected by Trizol reagent at indicated time points post infection. The negative strand genomic RNA of each sample was relatively quantified by RT-PCR and expressed as fold increase relative to that of sample collected at 0.5 hour post infection.