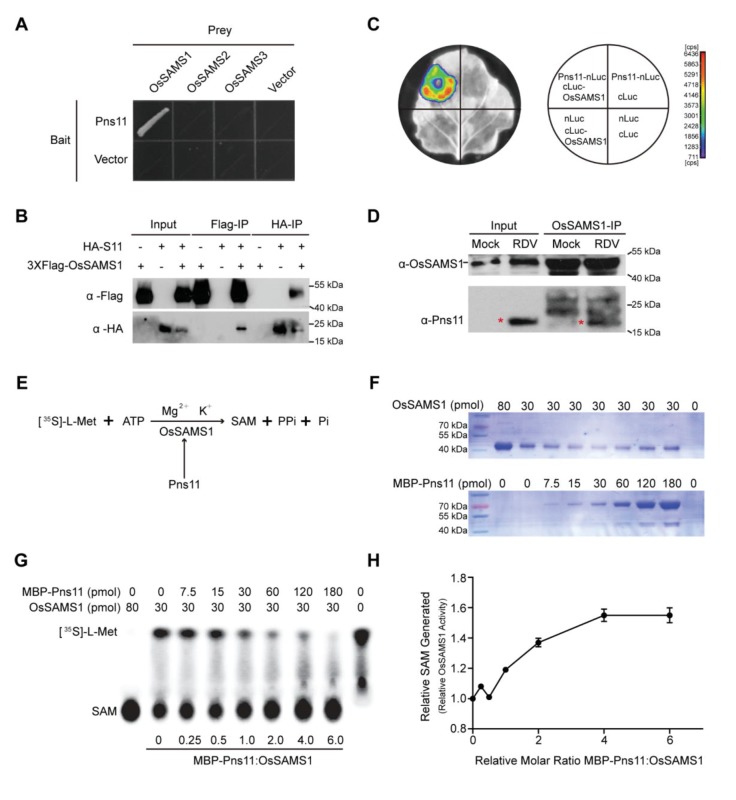
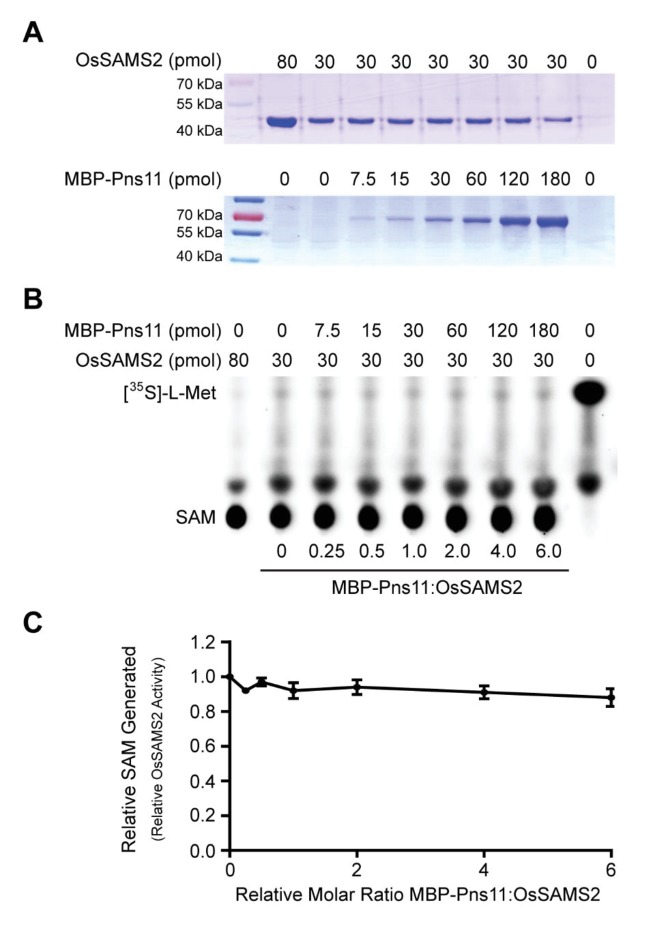
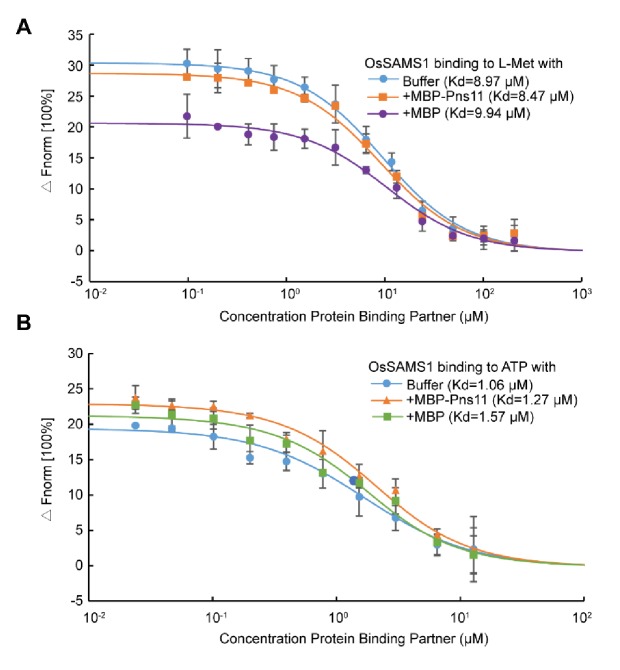
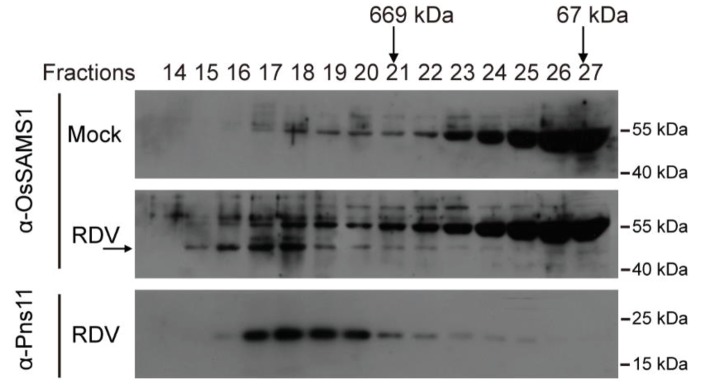
Figure 2. Pns11 interacts with OsSAMS1 and enhances its activity.
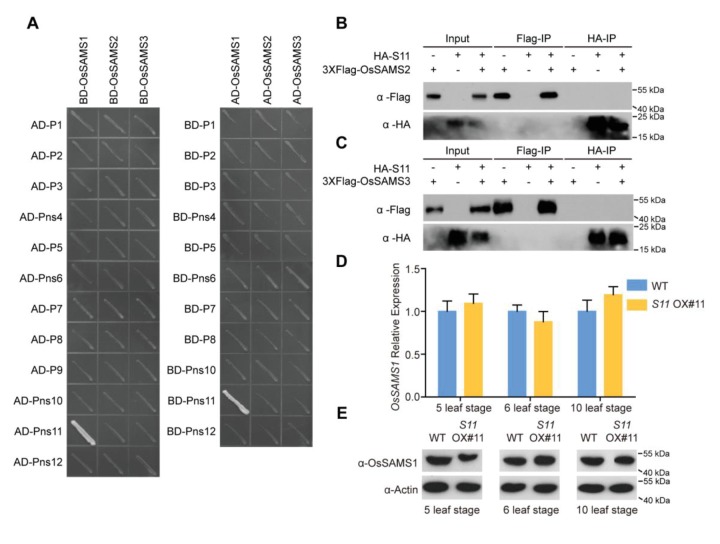
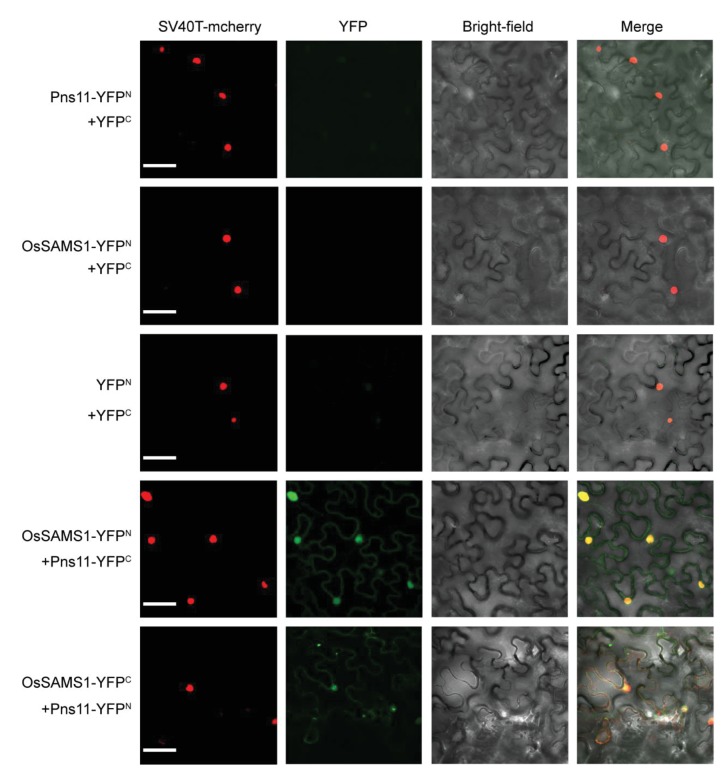
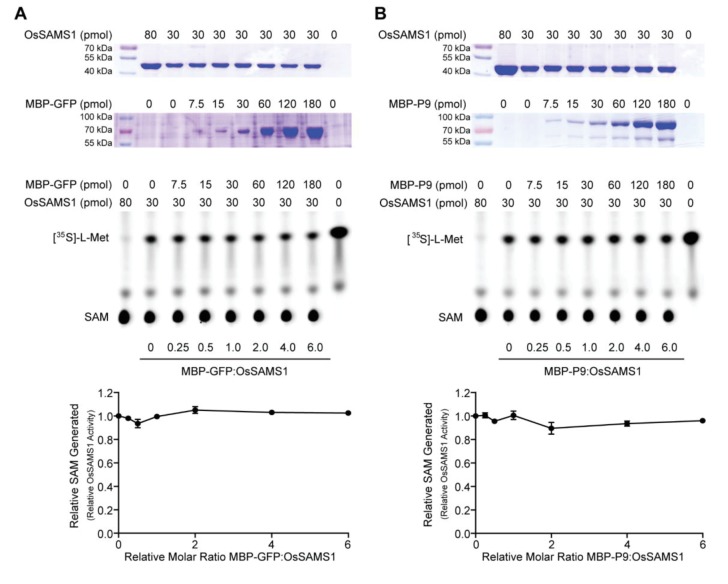
(A) Yeast two-hybrid assay for the interaction of Pns11 with OsSAMS1. The bait vector contained full-length Pns11; the prey vector contained OsSAMS1, OsSAMS2, or OsSAMS3. Yeast strains were cultured on the Trp –Leu –His –Ade selection medium. (B) Co-IP assay for the interaction of Pns11 with OsSAMS1. Pns11 and OsSAMS1 proteins were transiently expressed in Nicotiana benthamiana leaves for 3 days. Plant extracts were then immunoprecipitated using anti-Flag or anti-HA antibodies, separated on a 10% SDS-PAGE gel, and blotted with anti-Flag or anti-HA antibodies. (C) Luciferase complementation imaging assay for the interaction of Pns11 and OsSAMS1. Agrobacterium strain EHA105 harboring different construct combinations was infiltrated into different N. benthamiana leaf regions. After 3 days of infiltration, luciferase activities were recorded in these regions. Cps, signal counts per second. (D) In vivo pull-down assay confirmed the interaction between Pns11 and OsSAMS1 during RDV infection in rice using α-OsSAMS1 antibody. The red asterisks indicate the location of Pns11. Tissues were collected at 4 wpi. (E) Diagram of the assay. In the reaction, OsSAMS1 catalyzes a two-step reaction in the presence of Mg2+ and K+ that involves the transfer of the adenosyl moiety of ATP to methionine to form SAM and tripolyphosphate, which is subsequently cleaved to PPi and Pi. Conversion of L-[35S]-Met to SAM is activated by Pns11. (F) Coomassie brilliant blue staining of OsSAMS1 and Pns11 at the varying amounts of used in this assay. (G) Autoradiograph of a representative chromatogram showing SAM generated by OsSAMS1 in reactions containing varying molar ratios of maltose-binding protein (MBP)-Pns11 to OsSAMS1. The positions of labeled L-[35S]-Met substrate and SAM product are indicated. L-[35S]-Met and SAM in each reaction was calculated after phosphorimager quantitation of radioactivity in individual spots. (H) Stoichiometry of activation. The graph illustrates relative OsSAMS1 activity with increasing molar ratios of MBP-Pns11:OsSAMS1. Data were obtained from three independent experiments.