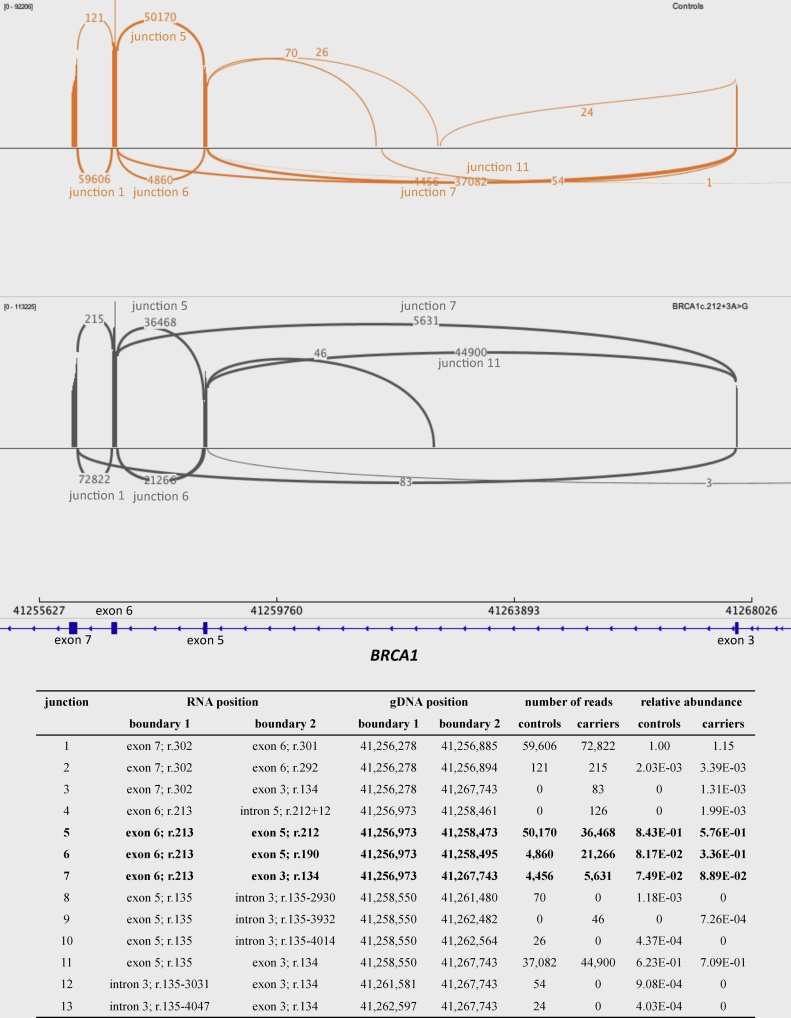
Fig. 1.
Sashimi plot of targeted RNA sequencing results. For the relative transcript quantification, the relative abundance of the three relevant transcripts was calculated as the number of reads covering the transcript-specific splice site divided by the total number of reads spanning the 3 splice sites (junctions 5–7). RNA positions are based on reference sequence NM_007294.3 starting at the A of the translation initiation codon (ATG) as position +1. Genomic positions are derived from hg19 reference sequence. 13 unique junctions were identified in controls (depicted in orange) and/or BRCA1 c.212+3A>G carriers (grey). At the boundary between exon 6 and 7 (junction 1), 59,606 and 72,822 reads were identified for the controls and the carriers respectively. As expected, three highly abundant transcripts are found on the other end of exon 6, connecting it to either exon 5 (junctions 5 and 6) or exon 3 (junction 7). For junction 5 (BRCA1-ex5FL), junction 6 (BRCA1-Δ22ntex5; c.191_212del22) and junction 7 (BRCA1-Δex5; c.135_212del78) the sum of the number of identified reads for these junctions is 59,486 and 69,525, which equals 99.80% and 87.01% of the number of reads at junction 1 for controls and the carriers respectively. In carriers the 13% difference is not explained by the novel junctions (junctions 3, 4 and 9), therefore we assume that the difference is caused by a percentage of prematurely terminated reads. The extremely low coverage also suggests that all junctions other than 1, 5, 6, 7 and 11 are artefacts, introduced by PCR or bioinformatics, rather than relevant isoforms. In BRCA1 c.212+3A>G carriers there is almost a five-fold relative difference in expression for BRCA1-Δ22ntex5 compared to controls, while BRCA1-Δex5 expression ratios are similar between carriers and controls.