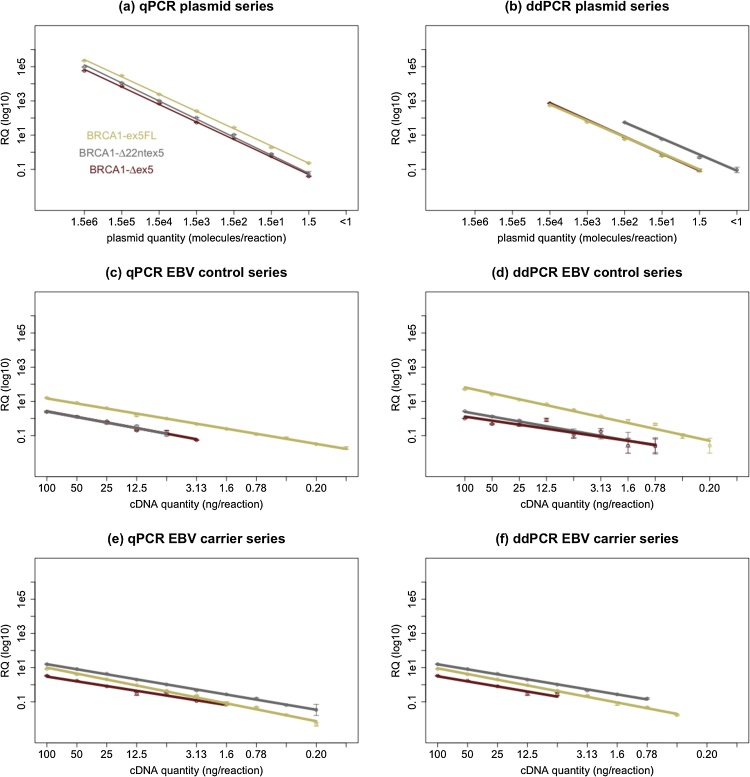
Fig. 2.
Linearity and dynamic range was tested on for all assays with both quantitative PCR methods, using dilution series containing transcript-specific plasmids ((a) and (b)) or cDNA from EBV samples (non-carriers ((c) and (d)) and carriers ((e) and (f)). Measured relative template quantity (RQ) is depicted on the y-axis in function of theoretical template concentration on the x-axis. Almost all assay-sample combinations show excellent linearity with both techniques (R2 > 0.98). Linearity was only worse for BRCA1-Δex5 in dilutions from EBV controls with ddPCR (R2 = 0.81). An important difference is that the dynamic range of qPCR across the dilution series was larger for several sample-target combinations, especially when quantifying transcript-specific plasmids. From these data we also estimated the LOB, LOD and LOQ values. For both methods LOB was equal to 0 (none of the no template controls yielded any amplification) and the LOQ and LOD for both methods reaches the theoretical minimum of 1 copy per reaction in transcript-specific plasmid dilution series. When using EBV derived samples the LOQ for both qPCR and ddPCR was found to range between 100 pg and 6.25 ng of cDNA depending on the abundance of a certain transcript in each sample.