Fig. 6.
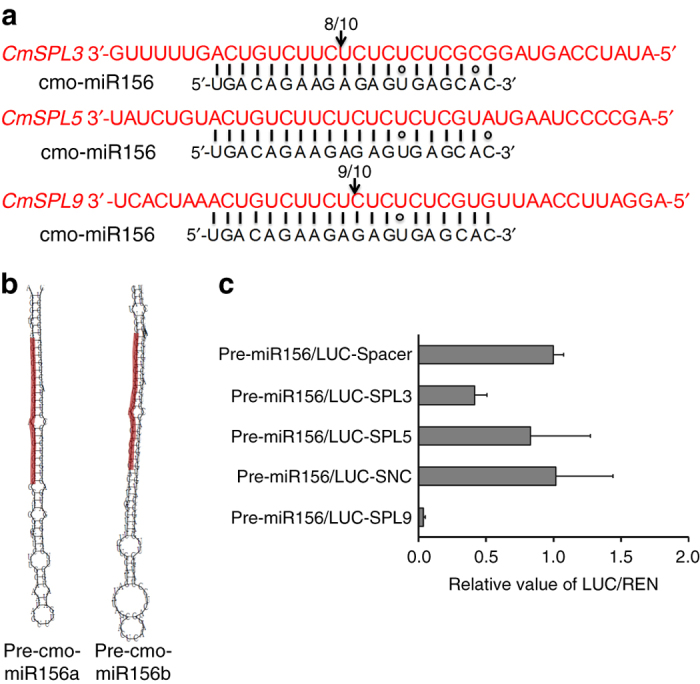
cmo-miR156 targets CmSPL3 and CmSPL9. a cmo-miR156-directed cleavage sites in the CmSPL genes were determined by 5′ RLM-RACE, and are highlighted by arrows. The number indicates the frequency of clones when validating the cleavage sites of the target mRNAs. b The precursor structures of cmo-miR156 were predicted by RNA structure software. miRNA sequences are highlighted in red. c A dual-luciferase reporter assay was used to quantitate cmo-miR156-mediated repression of target gene CmSPLs. 35S:cmo-miR156 precursor a (pre-miR156a) was co-transformed with CmSPL3 or 5 cleavage sites fused to the 3′UTR of the LUC gene, CmSPL9 cleavage site fused to the ORF of the LUC gene. LUC vectors carry a REN gene under the control of the 35S promoter as a positive control. Synonymous negative control (SNC) is a negative control with disruption of CmSPL9 cleavage site while maintaining identical amino acid sequence. Spacer is a 20 nt random sequence as negative control for CmSPL3 and CmSPL5. Nicotiana benthamiana leaves were infiltrated with the samples, and LUC and REN activities were assayed 3 days after infiltration. Three independent experiments were performed and error bars indicate standard deviation
