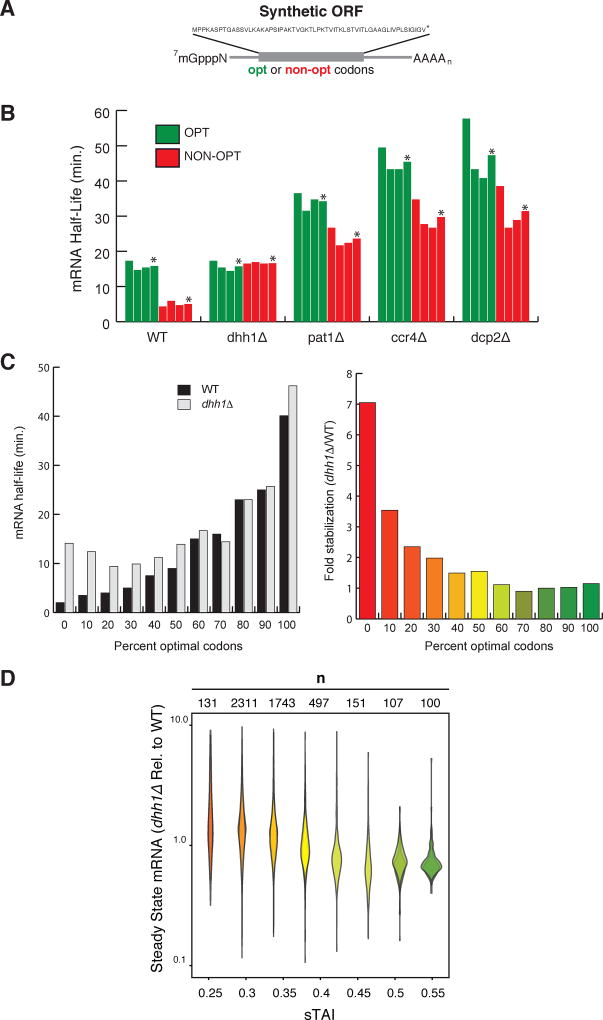
Figure 2. Dhh1p selectively stimulates the decay of mRNAs with low codon optimality.
See also Figure S2. (A) Representation of the synthetic mRNAs (SYN) and the encoded polypeptide sequence. Optimal (OPT) or non-optimal (NON-OPT) codons encoding the same peptide were used. The artificial peptide has no similarity to any known proteins. (B) The half-lives of SYN OPT and NON-OPT mRNAs in WT and different mutant strains were obtained from GAL1 shutoff experiments. Quantitations were normalized to the amount of SCR1 RNA. *Denotes average of 3 experiments. (C) Half-lives of HIS3 reporters from Figure 1B (GAL1 UAS constructs) in WT or dhh1Δ cells. Right panel indicates fold stabilization in a dhh1Δ cells vs. WT. (D) Quantification of steady state levels of mRNAs transcripts by RNA-Seq in dhh1Δ cells (RPKM) relative to WT cells (RPKM). mRNA transcripts are binned by sTAI, a numerical proxy for overall optimality. Shown are two biological replicates. A two-tailed Mann-Whitney test shows that low optimality mRNAs (sTAI = 0.25, Med. = 1.52) are enriched relative to high optimality mRNAs (sTAI = 0.55, Med. = 0.72) upon Dhh1p depletion, U = 1668, p < 2.2×10−16.