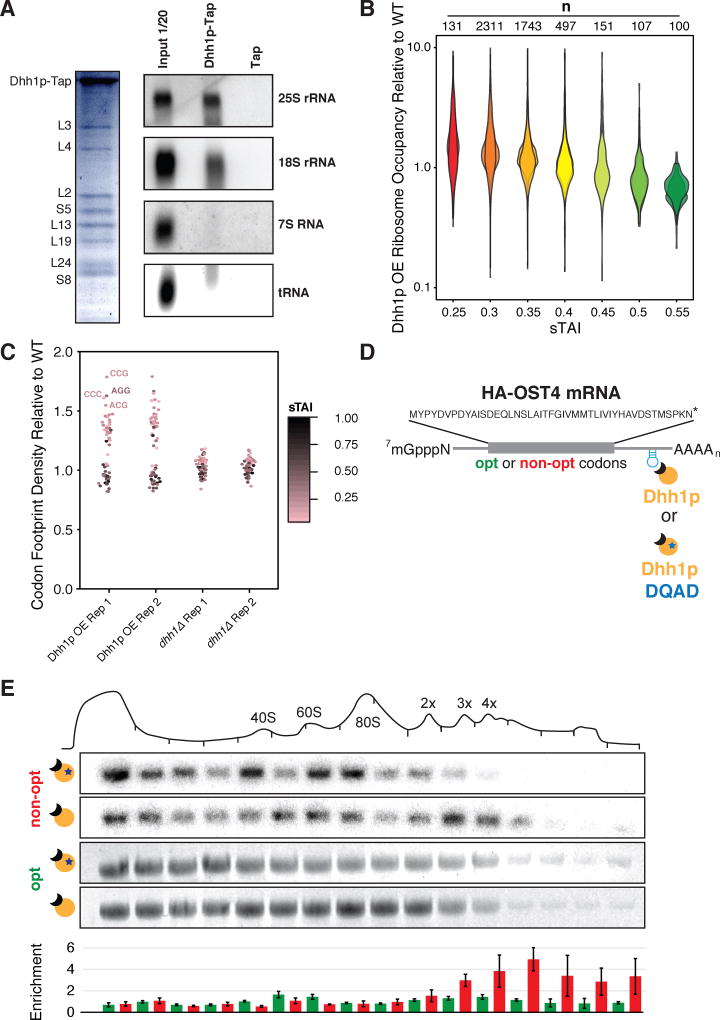
Figure 6. Dhh1p binds ribosomes and preferentially modulates ribosome occupancy on mRNAs with low codon optimality.
See also Figure S4. (A) Dhh1p-TAP purification followed by mass spectrometry (left, Coomassie blue gel staining) or Northern blots and specific probing for different rRNAs or tRNA (right). (B) Plotting the ribosome occupancy (average number of ribosomes per mRNA transcript) for mRNA transcripts under constitutive Dhh1p OE relative to WT conditions, binning transcripts by sTAI. Shown are two biological replicates. A two-tailed Mann-Whitney test shows that low optimality mRNAs (sTAI = 0.25, Med. = 1.30) have increased ribosome occupancy relative to high optimality mRNAs (sTAI = 0.55, Med. = 0.72), U = 1364, p < 2.2×10−16 upon Dhh1p overexpression (C) Quantifying the ribosome footprint density in the A-site under Dhh1p OE or dhh1Δ relative to WT. The identity of the codon in the A-site was determined by using 28-nt fragments as outlined previously (Ingolia et al., 2009). (D) Schematic of the reporter used in polysome occupancy assays. (E) Northern blots were used to quantify the enrichment (relative fractional occupancy) of optimal and non-optimal HA-OST4 mRNA along a polysome gradient upon tethering catalytically active and inactive Dhh1p. Reported values are averaged across three samples and presented with standard error. Shown are representative northern blots for the non-optimal and optimal mRNAs upon tethering of catalytically active and inactive Dhh1p.