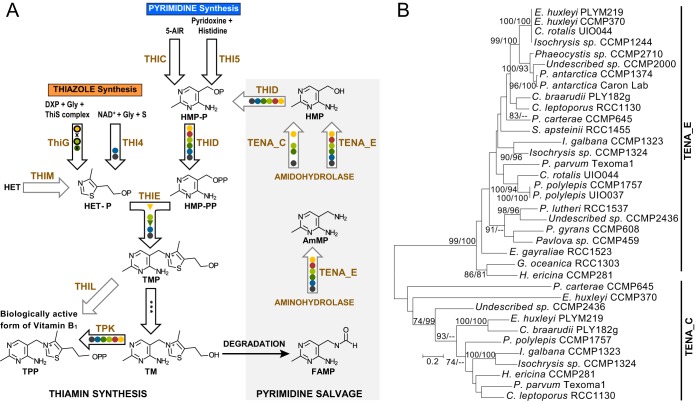
FIG 2 .
(A) Haptophyte thiamin biosynthesis and salvage enzymes. Enzymes (brown) and compounds (black) in the biosynthesis pathway based on reference 1 are shown with enzyme presence (solid circles) indicated for representatives of six haptophyte orders (colors as in Fig. 1); black outlines on circles indicate the gene is in the chloroplast genome, and triangles represent the presence of the THID and THIE fusion protein (TH1). Gray arrows show nonessential or salvage-related pathway steps (also with gray background, right side of panel). Haptophyte orders for which chloroplast genomes are not available are indicated by “x.” Unspecific phosphatases are represented by *. 5-AIR, 5-aminoimidazole ribotide; DXP, 1-deoxy-d-xylulose 5-phosphate; Gly, glycine; NAD+, NAD; S, reduced sulfur species; TM, thiamin; P, phosphate group; PP, pyrophosphate; HET, 4-methyl-5-β-hydroxyethylthiazole; HMP, 4-amino-5-hydroxymethyl-2-methylpyrimidine; AmMP, 4-amino-5-aminomethyl-2-methylpyrimidine; FAMP, N-formyl-4-amino-5-(aminomethyl)-2-methylpyrimidine. (B) Unrooted maximum likelihood phylogeny of haptophyte TENA_E and TENA_C proteins based on 390 amino acid positions. Bootstrap support (100 replicates) is shown in the order maximum likelihood/neighbor joining for values of ≥70%.