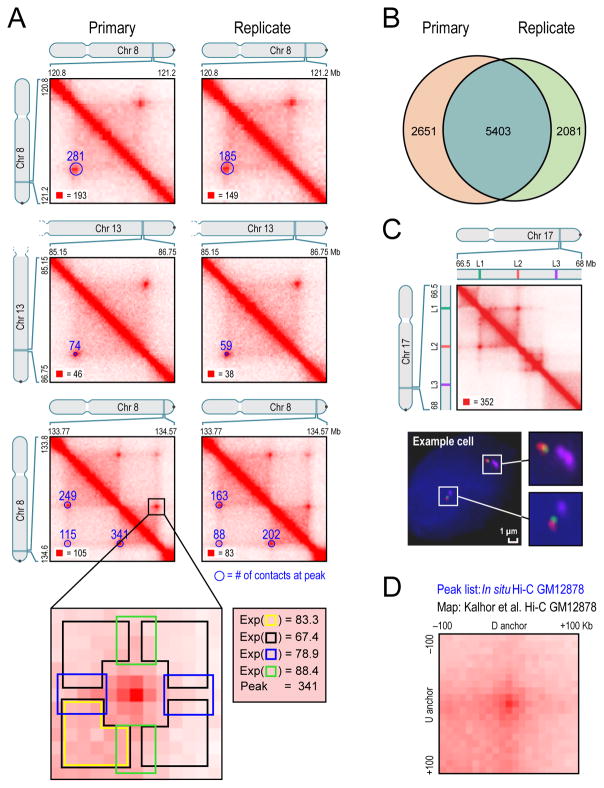
Fig. 3. We identify thousands of chromatin loops genome-wide using a local background model.
(A) We identify peaks by detecting pixels that are enriched with respect to four local neighborhoods (blowout): horizontal (blue), vertical (green), lower-left (yellow), and donut (black). These “peak” pixels are marked with blue circles (radius=20Kb) in the lower-left of each heatmap. The number of raw contacts at each peak is indicated. Left: primary GM12878 map; Right: replicate; annotations are completely independent. All contact matrices in these figures are 10Kb resolution unless noted. (B) Overlap between replicates. (C) (Top) Location of 3D-FISH probes (Bottom) Example cell. (D) APA plot shows the aggregate signal from the 9948 GM12878 loops we report by summing submatrices surrounding each peak in a low-resolution GM12878 Hi-C map due to Kalhor et al. See also Figure S4, Table S3, Table S4, and Table S5.