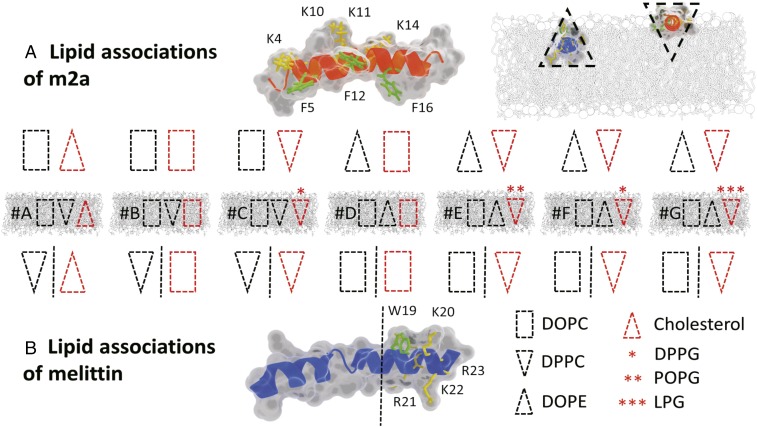
Fig. 2.
The lipid associations of melittin and m2a in the membrane systems A–G, with lipid topology indicated by the block shapes. A red outline indicates lipids with a clustering mechanism either via electrostatics for the anionic lipids or via aromatic π-stacking with phenylalanine (F) and tryptophan (W) for cholesterol. (A) The block shapes indicate the topology of the lipids expected to associate with m2a. (B) The lipid topologies expected to associate with melittin. Shapes to the left of the dashed line associate with the peptides’ helical face, while those to the right cluster with the peptides’ tryptophan and cationic residues at the C terminus. (A, Top Right, Inset) The deeper penetration of melittin into the outer leaflet allows it to act as a negative curvature membrane wedge, while the shallower penetration of m2a results in a positive curvature wedge effect indicated by the appropriate block shapes.