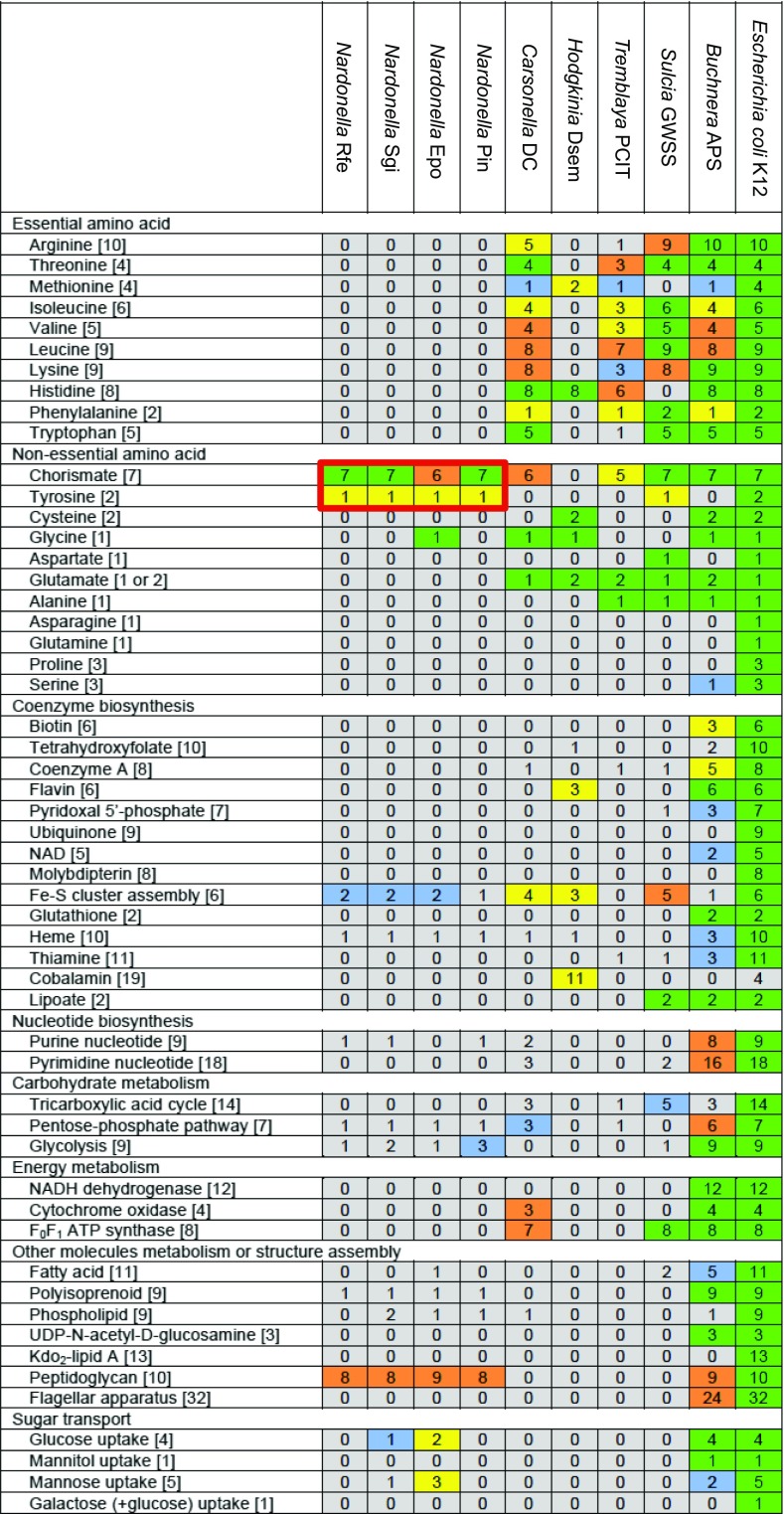
Fig. 3.
Comparison of the metabolic gene repertoire between Nardonella genomes and other extremely reduced symbiont genomes. The minimal number of genes for a metabolic pathway is shown in each of the brackets. Each color indicates the ratio of retained genes to the minimal gene set for a metabolic pathway: green for 100%, orange for 75–99%, yellow for 50–74%, blue for 25–49%, and gray for 0–24%. Nardonella’s tyrosine synthesis pathway genes are highlighted in red. In the Nardonella Epo genome, aroE gene, located between def and sufE genes in the other Nardonella genomes, is lost and replaced by a 168-bp spacer sequence, which may be relevant to the fact that its host E. postfasciatus is smaller in size with thinner cuticle in comparison with the other large and hard weevil species R. ferrugineus, S. gigas, and P. infernalis.