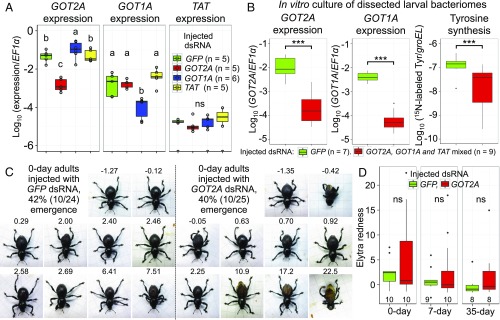
Fig. 9.
RNA interference (RNAi) suppression of host’s tyrosine synthesis genes up-regulated in the bacteriome of P. infernalis, and its effects on cuticle formation and pigmentation. (A) RNAi targeting GOT2, GOT1, and TAT by injecting double-stranded RNA (dsRNA) into mature larvae. Expression levels of GOT2A, GOT1A, and TAT were measured by quantitative RT-PCR 3 d after the injection. Different letters (a, b, c) indicate statistically significant differences (likelihood-ratio test of GLM and post hoc multiple comparisons; P < 0.05). (B) Tyrosine synthesis activity of dissected larval bacteriomes suppressed by RNAi of GOT2 and GOT1. Mature larvae were injected with dsRNAs, their bacteriomes were dissected 7 d after the injection and cultured in a medium containing 15N-labeled glutamine for 2 h, and the bacteriomes were subjected to quantitative RT-PCR, whereas the culture media were analyzed by LC-MS for quantification of 15N-labeled tyrosine. Asterisks indicate statistically significant differences (likelihood-ratio test of GLM assuming a gamma error distribution; ***P < 0.001). (C) Images of adult insects on the day of emergence, which were subjected to larval injection with either GFP dsRNA or GOT2A dsRNA. Numbers on the top of the images indicate the values of elytra redness. (D) Comparison of elytra redness between the adult insects 0, 7, and 35 d after emergence. “ns” indicates no statistically significant difference (Wilcoxon rank sum test; P > 0.05). Tukey box plots are as shown in Fig. 4 G and H. Note that the primers for quantitative RT-PCR are highly specific, whereas dsRNAs for RNAi may potentially cause some cross-suppressions: dsRNA for GOT2A may also recognize GOT2B, and dsRNA for GOT1A may also target GOT1B and GOT1C (SI Appendix, Fig. S8).